Six known CDC genes [6], line 2. Their coefficients of variation, shown on line 1. Each are highly correlated, line 3, to the genes of line 4; each of which have the higher coefficients of variation, shown on line 5.
Abstract
A novel approach to the dynamics of gene assemblies is presented. Central concepts are high-value genes; correlated activity; orderly unfolding of gene dynamics; dynamic mode decomposition; DMD unraveling dynamics. This is carried out for the Orlando et al. yeast database. It is shown that the yeast cell division cycle, CDC, only requires a six-dimensional space, formed by three complex temporal modal pairs: (1) a fast clock mother cohort; (2) a slower clock daughter cell cohort; and (3) an unrelated inherent gene expression. A derived set of sixty high-value genes serves as a model for the correlated unfolding of gene activity. Confirmation of this choice comes from an independent database and other considerations. The present analysis leads to a Fourier description, for the very sparsely sampled laboratory data. From this, resolved peak times of gene expression are obtained. This in turn leads to precise times of expression in the unfolding of the CDC genes. The activation of each gene appears as uncoupled dynamics originating in the mother and daughter cohorts, and of different durations. This leads to estimates of the composition of the original laboratory data. A theory-based yeast modeling framework is proposed, and additionally new experiments are suggested.
Keywords
- cell division cycle
- co-regulated genes
- high-value genes
- dynamic mode decomposition
- low dimensional models
1. Introduction
The blueprint of a life form is contained in its DNA genome, as formed from the four bases, [A, C, G, T], as assembled in the double helix [1]. The genome contains instructions for decoding itself, constructing itself, duplicating itself, and inserting these instructions in the duplicate. This
Budding yeast, (Saccharomyces)
To avoid misunderstanding, the present goal is the dynamic unfolding of gene dynamics in contrast to the dynamics of individual genes, as for example in [10].
2. Yeast
The fate of a budding yeast cell is to divide asymmetrically into a mother and daughter cell. The cell period is in the range of an hour or two. In their pioneering paper Spellman et al. [8] describe several different means by which to assemble a population of quasi-identical daughter cells, for purposes of tracking the dynamics of the CDC. As in [6] elutriation will be used here to assemble a population of mother and daughter cells. Laboratory data monitors 5716 genes by sampling, at 16-min intervals, 15 times, covering roughly 2 cycles of the CDC. The database contains two wild type sets, WT1 and WT2 (referred to here as G1 and G2, and two mutant sets). Each experimental dataset is thus represented by a matrix of 5716 rows and 15 columns, that describes the temporal expression of all genes, denoted by,
where
where
For our purposes, instead of (1) the mean subtracted form,
is an effective beginning.
where
One can reasonably conclude from Figure 1, that the analysis can be reduced to the six modes, ahead of the
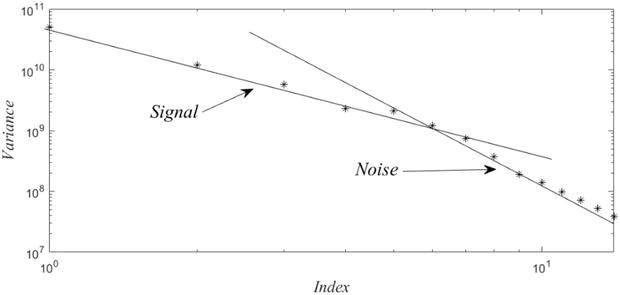
Figure 1.
Log-log plots of the variances reveal that these follow two different power laws, which by routine arguments can be associated with signal or noise.
where
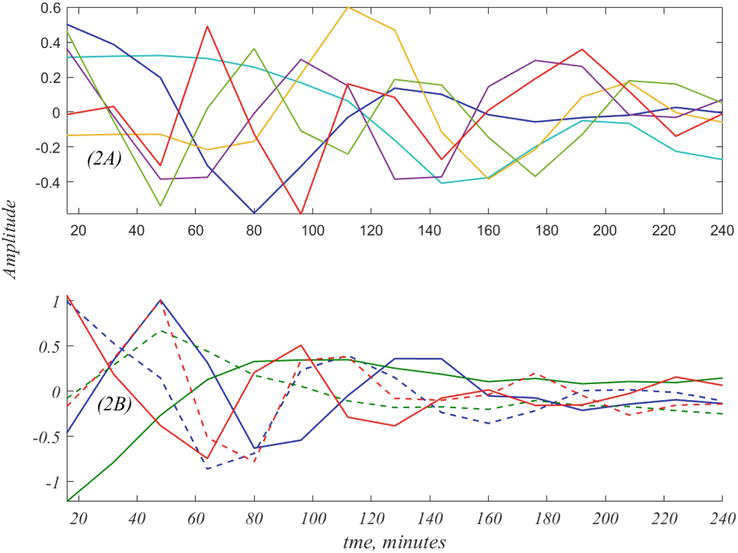
Figure 2.
The six time histories of
The plots of Figure 2(A) may be regarded as artifactual consequences of SVD, that only hint at the true dynamics. The goal of DMD is to recast the data in terms of the true exponential frequencies that are latent in the data. Thus, the curves of Figure 2(A) are entangled versions of the true dynamics, shown in Figure 2(B). In Section 8 we obtain the 6 × 6 matrix,
provides a generalization of SVD, (4), that is appropriate for data that have a rational dynamic ordering of columns.
3. High value genes
One can reasonably anticipate two limiting forms of gene expression; steady expression, as might be the case, for the proteins that form the cell wall and membrane; and a briefer activation and later inactivation on a shorter timescale, as might be the case for formation of the cell nucleus and its components. As a criterion for distinguishing these two limits consider the coefficient of variation,
where
| CV | 0.1845 | 0.7216 | 0.3936 | 0.2337 | 0.6861 | 0.2903 |
| Gene | YDL155W | YGR108W | YGR109C | YLR210W | YPR119W | YPR120C |
| CorrCoef | 0.9843 | 0.9878 | 1 | 1 | 0.9950 | 0.9864 |
| Gene | YMR144W | YGL021W | YGR109C | YLR210W | YGL021W | YJL181W |
| CV | 0.3475 | 0.7222 | 0.3936 | 0.2337 | 0.7222 | 0.3128 |
Table 1.
Co-regulation implies correlated activity. The fourth line of the table shows genes highly correlated to those of the second row, shown on the third line, to the genes of the second line. The implication of the Table is that the genes of the fourth line are better gene representatives. Peak times of two like genes are virtually identical.
In general, any gene can be well correlated to many genes. This is illustrated in the next figure the for genes correlated with GR108W, with gene names and coefficients of variation in the legend. YGL021W with a CV = 0.7222 the best exemplar of this set of highly correlated genes.
As is clear from Figure 3, peak locations, ipso facto, must occur at sampling locations, but would be better resolved by interpolation.
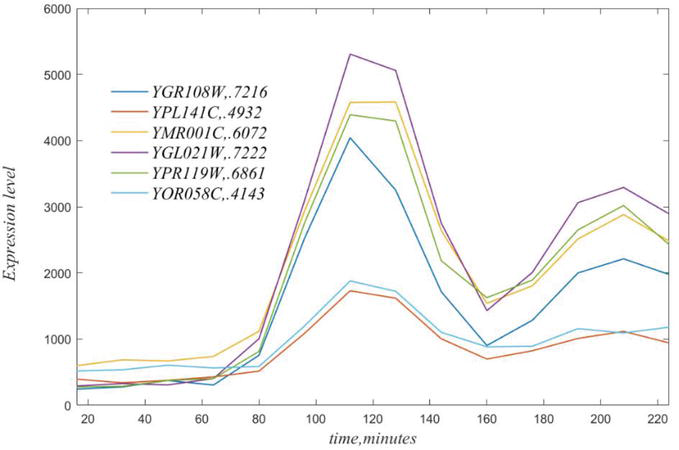
Figure 3.
Time courses of genes highly correlated to YGR108W, see legend. Note that each of these has a peak at
3.1 Gene selection
As mentioned above there are 1192 genes for which CV > 0.25. These will be regarded as a starting point for selecting high value genes. A large number of traces exhibit pure exponential decay, starting at extremely high expression values, and are regarded as artefactual. Thus, a second criterion is restriction to time traces that start with relatively small expression, as is the case in Figure 3. For example, a restriction to initial value of a gene at 16 min, of <450, results in a well-correlated set of 109 genes. Figure 4 shows the correlation image based on the correlation criterion,
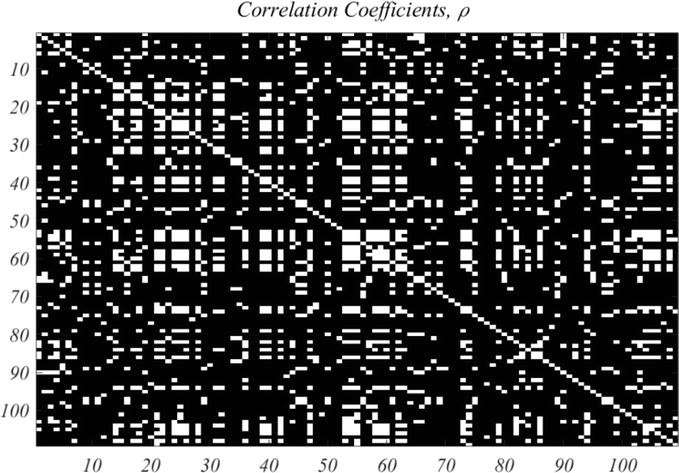
Figure 4.
Correlation coefficient of the 109 selected genes, with correlation coefficient,
The above figure is based on
where
4. Analysis of the high value genes
The DMD analysis of
where
This form speaks volumes. Since each
where
And enduring puzzle of SVD analysis has been the origin of the time courses of
4.1 Unfolding the CDC
The goal is acquisition of a data-based model of the CDC, under the assumption that CDC is a temporal unfolding of genes, with defined activation times. As is clear from Figure 3, time resolution is limited by course sample times. The exponential representation of (13) induces a natural Fourier representation that overcomes this limitation. From the calculated frequencies, (9), we can
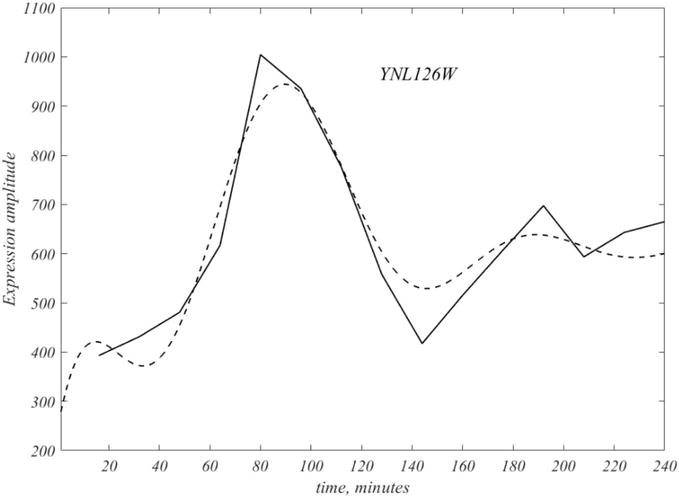
Figure 5.
The dashed curve represents the Fourier interpolated version of the original polygonal data of the gene YNL126W, and is the first of the 109 genes. Note the interpolation covers the initial 16 min, and also runs over the second attenuated cycle.
Inspection of the 109 highly sampled genes reveals that 43 take on negative values, and 8 have a peak at
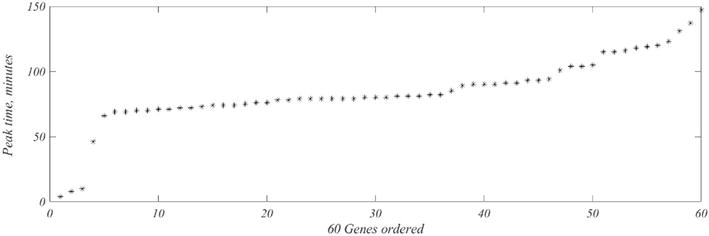
Figure 6.
Times,
In Figure 7, the left image shows the trajectory of gene expression for peak times arranged in ascending order. This compares favorably with the phase plots that appear in [6]. At the right is the comparable plot for WT2 under the same gene ordering. It should be noted that the Orlando et al. plots are based on their 440
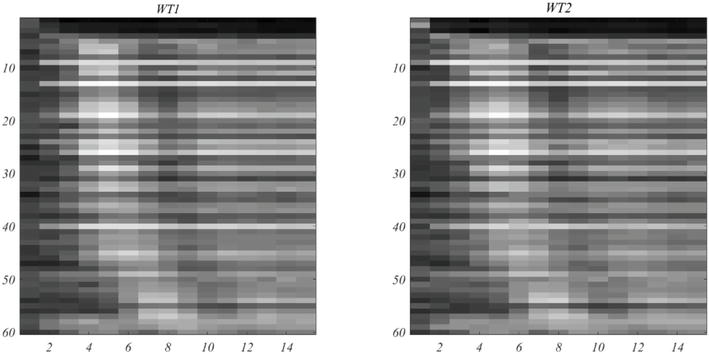
Figure 7.
Track of mRNA expression of the 60 selected genes,
5. Coherent gene sets
Here,
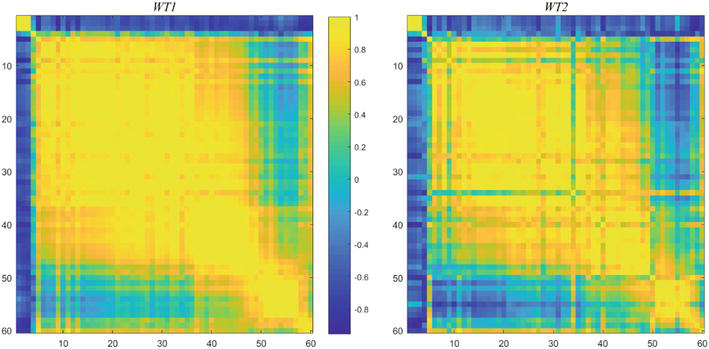
Figure 8.
Correlation coefficients,
The figure on the right is the result of the same calculation, based on the selected ordering applied to WT2. This provides a compelling demonstration that the 60 choice genes are “strongly co-regulated”, in general. A much larger set, 413 genes, similarly constructed, produces an intersection of 204 genes with the Spellman co-regulated set of 800.
Unfolding times is regarded as a reasonable hypothesis for gene ordering; though other possibilities may be considered. For example, ordering the genes in terms of descending correlations,
6. The single yeast cell
Figure 3, exhibits a typical gene time course, and displays single gene expression duration over many tens of minutes. However, the accepted estimate for the duration of gene transcription and translation is 1–2 min [15], and for convenience this value is taken to be 1 min in the calculations performed below. To explain what might appear to be an inconsistency, we review the data acquisition procedure, and as will be seen is due to the different maturation periods of mother and daughter cells, and randomness.
In experiments, after assembly of a suitable pool of yeast cells, aliquot removals along with genetic snapshots are obtained at 16 min intervals, and repeated 15 times. The result is the report of mRNA expression for each gene at each sampling instant. According to [6] each sampling, contains more than 200 yeast cells. To obtain a sense of the process consider gene YGR174-A, the earliest activated gene, of the 60 gene set, as deconstructed in Figure 9.
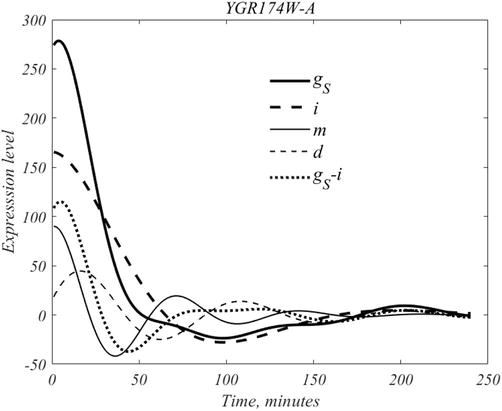
Figure 9.
The dynamical composition of gene
The mean subtracted form of this genes is denoted by gS; DMD produces the four traces related by,
where
The mother cohort, m, has a peak of 90 at
follows. Thus, there are more than three times as many daughter cells as mother cells in an average aliquot.
For formal purposes the inherent signal and the mean will be divided into 2 parts, as follows,
so that the total signal is equals
where mother, daughter, inherent and mean our shown in Figure 9. As shown in Figure 10, the full signal this gene equals
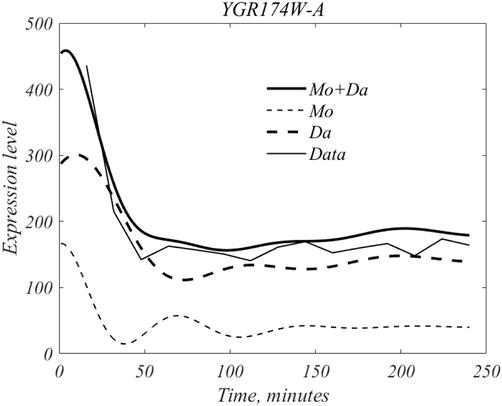
Figure 10.
Mother cell dynamics are initiated at roughly
To summarize, genes are expressed both in a background manner, inherent expression, and in a scheduled manner, to peak at some specific time. It is also clear from this analysis that the activities of mother and daughter cells are not coupled.
6.1 A yeast model
Next, we consider a proposed computational model of the yeast cell. For this we focus on the gene traces displayed in Figure 10. While additional genes might be included in the model, the uncertainties in experimental results [15, 16] do not justify such generalizations. Our purpose in this exercise is merely to demonstrates that a practical framework can be created.
To start, it is noted that estimates of protein molecules per yeast cell are in the range of ∼
The ratio pD/pM = .26 is remarkably close to the above mentioned
On the basis of these deliberations one might contemplate creating an algorithmic model of the CDC. Randomization can be introduced through variations in mother and daughter CDC periods, and variations in the number of mother and daughter cells, say adding up to roughly 200. This is a future project, which can useful only with better knowledge and precision of the quantities involved.
7. Additional comments
The high degree of correlation, seen in Figure 6 tells little beyond timing. For example, it does not imply anything certain about gene interactions, nor is there any information about the activation and deactivation times of genes. The mechanism by which budding yeast cell assembles itself, is an open question. Since no outside intervention is in play, it is noncontroversial to presume, that the cell self-assembles. Just how this self-assembly takes place is another open question, it might e.g. only be a matter of proteins falling into their proper place, based on the timing of gene expression. In this case the cell model is an
In an effort to introduce some additional theory it is noted activation and inactivation of gene expression may be likened to an equilibrium disturbance, followed by restoration, (suggestive of wave phenomena.). In this connection it is noted that the mother and daughter modes that describe the CDC each follow oscillator dynamics [9]
which has solutions in the form,
In this connection, we can define a new variable,
which since
The pair of Eqs. (20) and (21) may be viewed as a coarse-grained version of the
A key result of the present investigation is the remarkable ability of DMD, to distinguish dynamic characteristics of the mother and daughter cohorts. Gene experiments typically attempt to sequester daughter cohorts, and it is natural on the basis of the methods pursued here to consider what the outcome would have been of considering complementary populations, and furthermore to monitor the base population, without any form of sequestering. Given the remarkable ability of DMD to parse dynamic activity more complicated population yeast populations might be considered. Hopefully, this ability to distinguish yeast subpopulations will lead to new ways to probe into the yeast life form. Another future goal, is that the algorithmic model touched on here can be further advanced, since a falsifiable model is always desirable.
8. Methods
8.1 Dynamic mode decomposition
Traditional signal analysis is based on the hypothesis that a
with corresponding complex signal,
Typically, a laboratory signal is a uniformly sampled version of the continuous case. Suppose for example the uniformly sampled times are,
in which case (23) becomes the geometric sequence.
where
which is therefore the generator of the sampled signal. Reciprocally, if
which can be viewed as a case of many equations for the one unknown,
and the (least-squares) minimization of (28) produces the solution
in which terms (29) can be written as
for later purposes observe that if the problem is posed as,
then it is solved by
where
In the general case, of multiple complex signals, we are confronted by a matrix
with
so that
which is solved by the Moore-Penrose inverse,
The spectral decomposition of the generator matrix,
produce complex frequencies as eigenvalues of the diagonal matrix
where
which is just the inverse of (26).
In the interest of brevity, we forgo examples. Consideration of synthetic data generated by,
with each of the M trials an admixture of m complex signals and randomly chosen coefficients shows a remarkable accuracy in recovering the frequency content after relatively few trials.
9. Conclusions
This paper represents a substantial extension of an earlier preliminary analysis [9] of the high quality yeast database assembled by Orlando et al. [6].
The principal focus of this paper is the introduction of advanced mathematical methods that should be useful under a wider set of circumstances. For example, for general dynamical molecular biological data sets, and for when better resolved data becomes available for
The Singular Value Decomposition, SVD, is the chosen mathematical framework for dealing with the dynamical structure of the yeast data and is believed likely to play an important role in examining dynamical biological data of a general nature. A shortcoming of SVD is that the dynamics generated by SVD is severely constrained by the underlying methodology of SVD. This shortcoming was repaired by Schmidt [13], by a method that is termed dynamic mode decomposition, DMD. This is fully treated in Section 8 of this paper, in particular see (13). In brief, the result is that the CDC is well approximated by a single mode that depicts the dynamics in terms of timescales representative of mother cells, and daughter cells.
It is an opinion that future more highly sampled data lead to the same qualitative description that is more refined, and accurate.
References
- 1.
Watson JD, Crick FH. Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Nature. (Wiley, New York). 1953; 171 (4356):737-738 - 2.
Neumann JV. The General and Logical Theory of Automata. Vol. 1951. New York: Wiley; 1951. pp. 1-41 - 3.
Schrödinger E. What is Life?: With Mind and Matter and Autobiographical Sketches. Cambridge University Press; 1992 - 4.
Nirenberg MW, Matthaei JH. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proceedings of the National Academy of Sciences. 1961;47 (10):1588-1602 - 5.
Crick F. Central dogma of molecular biology. Nature. 1970; 227 (5258):561-563 - 6.
Orlando DA et al. Global control of cell-cycle transcription by coupled CDK and network oscillators. Nature. 2008; 453 (7197):944-947 - 7.
Cho RJ et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Molecular Cell. 1998; 2 (1):65-73 - 8.
Spellman PT et al. Comprehensive identification of cell cycle–regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Molecular Biology of the Cell. 1998;9 (12):3273-3297 - 9.
Sirovich L. A novel analysis of gene array data: Yeast cell cycle. Biology Methods and Protocols. 2020; 5 (1)1-10 - 10.
Tyson JJ. Modeling the cell division cycle: cdc2 and cyclin interactions. Proceedings of the National Academy of Sciences. 1991; 88 (16):7328-7332 - 11.
Sirovich L. Turbulence and the dynamics of coherent structures. I. Coherent structures. Quarterly of Applied Mathematics. 1987; 45 (3):561-571 - 12.
Lax PD. Linear Algebra and its Applications. New York: Wiley; 2007. p. 2007 - 13.
Schmid PJ. Dynamic mode decomposition of numerical and experimental data. Journal of Fluid Mechanics. 2010; 656 :5-28 - 14.
Kutz JN et al. Dynamic Mode Decomposition: Data-Driven Modeling of Complex Systems. Philadelphia: SIAM; 2016 - 15.
Milo R, Phillips R. Cell Biology by the Numbers. New York: Garland Science; 2015 - 16.
Gerstein MB et al. What is a gene, post-ENCODE? History and updated definition. Genome Research. 2007; 17 (6):669-681 - 17.
Beadle GW, Tatum EL. Genetic control of biochemical reactions in Neurospora. Proceedings of the National Academy of Sciences of the United States of America. 1941; 27 (11):499 - 18.
Golub GH, Van Loan CF. Matrix Computation. 1989. Johns Hopkins. Baltimore, MD: University Press; 1989 - 19.
Nyquist H. Certain topics in telegraph transmission theory. Transactions of the American Institute of Electrical Engineers. 1928; 47 (2):617-644