Analytical methods used for salivary analysis.
Abstract
Oral cancer carcinogenesis is a complex process that outlines the implication of multiple mechanisms that lead to the development of this specific malignancy. The high heterogeneity of this disease is a key factor that controls the progression and treatment response, influencing the survival rate. The multifactorial etiology, the genetic alterations and the diagnosis in advanced stages are directly involved in the high mortality rate of this pathology. Currently, the gold standard for oral cancer diagnosis is represented by the tissue biopsy and its histopathological examination, procedure that in time revealed several disadvantages. Recent research focused on a non-invasive, fast and reliable diagnosis approach based on the use of saliva. Saliva through its components provides information regarding specific salivary molecules, proteomic and genomic changes linked to oral cancer occurrence and progression. By using saliva as a diagnosis tool, it offers an important perspective of the tumor environment, designing a complete molecular profile of the tumor by creating the concept of personalized medicine.
Keywords
- oral cancer
- early diagnosis
- non-invasive
- saliva
- oral squamous cell carcinoma
- oral cancer screening
- salivaomics
1. Introduction
Oral cancer is currently an arising healthcare problem worldwide, challenging researchers in improving the prevention strategies and early diagnosis. The incidence in the last years determined oral cancer to represent 12% out of all the encountered malignancies [1]. Approximately, 90% of all the oral cancer cases arise from the oral mucosa lining and are represented by oral squamous cell carcinoma (OSCC) [2]. The diagnosis of oral malignancy implies a transition from a normal oral mucosa to the occurrence of benign hyperplasia, followed by dysplasia that left untreated is the precursor of carcinoma
One of the main concerns regarding this malignancy is the high mortality and morbidity, mostly due to a late diagnosis in advanced stages. Compared to the other types of cancers, oral cancer could benefit from an early diagnosis due to the direct visibility of the oral mucosa and the possibility of a proper examination identifying potential existing premalignant lesions.
It is considered to be a multifactorial disease, emerging from a complex interaction between the epigenetic and genetic factors, with important cellular and molecular interactions [4]. Despite the advancements made in the diagnosis field, the survival rate associated with oral cancer is only 50%, mainly due to the late presentation of the patients for a diagnosis [5]. Currently, besides a good oral examination, several chair-side investigations and laboratory procedures, the high-end diagnostic approaches that include procedures such as nano-diagnostics, the analysis of biofluids, liquid biopsy, genomics, proteomics and metabolomics technology, offer a new perspective for screening and early diagnosis [6].
Currently, the detection of oral cancer is made through the conventional oral examination (COE) and further by performing a biopsy and a histopathological examination of the tissue sample. Until present, the biopsy is considered to be the gold standard for the diagnosis of oral malignancies, although it is an invasive procedure that often causes unwanted consequences for the patient. In order to reduce the unnecessary biopsy cases and to implement a strategy for the early diagnosis of oral cancer, it is important to understand the complex molecular pathways of carcinogenesis that occur at an early stage and are based on the identification of multiple genetic, epigenetic, proteomic and metabolomic biomarkers. The quantification of these markers represents the start-point of a new diagnosis era that targets the non-invasive approaches. Introducing the use of biofluids such as blood and saliva brought additional information regarding the screening, early diagnosis and monitoring of oral diseases.
Recently, the use of saliva as an oral cancer diagnosis tool revealed promising results. Taking into consideration the permanent contact of this fluid to the tumoral microenvironment, a direct release by the tumor of multiple potential biomarkers and the advantage of offering a personalized perspective, has transformed saliva into an appealing fluid that could be successfully introduced in the diagnosis field. Research showed that specific biomarkers signatures present in the saliva of oral cancer patients could be identified through a non-invasive approach, having a high sensitivity and specificity, opening a new path toward the personalized medicine concept. The progress that has been made by introducing the salivary diagnosis and understanding the complex characteristics of saliva, through a simple, non-invasive and cost-effective collection method, offers clinicians and patients an important asset in the diagnosis and treatment steps.
2. Saliva: an appealing biofluid
2.1 Saliva: generalities, composition and function
Saliva is produced and excreted by the three major salivary glands (submandibular, parotid and sublingual) and the minor salivary glands that are localized in multiple areas of the oral mucosa [7]. Salivary secretion can be influenced by several physiological and pathological conditions that determine the quantitative and qualitative changes in its composition. The average amount of saliva produced by healthy adults is between 500 and 1000 mL saliva/day, with an average salivary flow rate of 0.3–0.4 mL/min [8]. The increase of the salivary flow can be influenced by oral hygiene, exercise, age, associated drugs, external stimuli such as smell and taste and hormonal changes [9]. Each salivary gland secrets specific saliva, a serous or a mucous type, and its collection can be performed either directly from a specific gland or as the collection of whole mouth saliva, representing the total amount of saliva excreted by all the salivary glands (Figure 1).
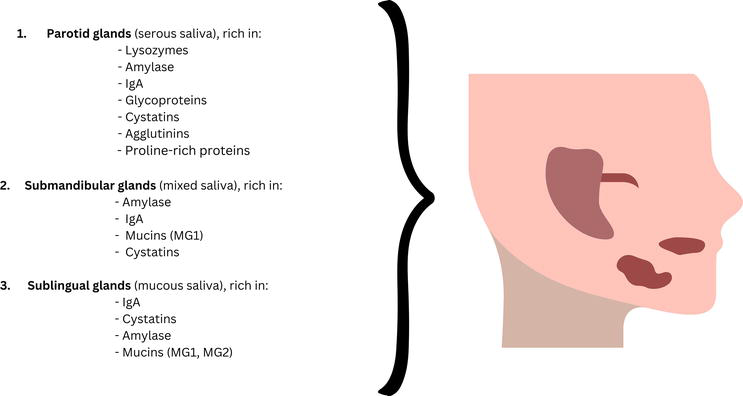
Figure 1.
Major salivary glands—Type of saliva and its composition.
Saliva consists of 99.5% water, 0.2% inorganic material and enzymes, 0.3% being represented by proteins. Its secretion is controlled by the sympathetic and parasympathetic nerves [10].
The salivary composition varies individually and in relationship with the circadian rhythm. The inorganic composition of saliva is represented by calcium, phosphate, potassium, sodium, chloride, bicarbonate, magnesium and thiocyanate. The organic composition consists of components such as immunoglobulins, mucins, uric acid, lactoferrin, cytokines, hormones and enzymes (amylase, lipase, peroxidase and lysozyme) [10]. All these constituents are secreted by the acinar cells in the salivary glands and afterward released into the oral cavity. The electrolyte composition secreted by the acinar cells is similar to the electrolyte composition of the ultrafiltrate of plasma, outlining its important diagnosis potential [11].
Among the composition represented by the molecules that are present in the salivary glands, the gingival crevicular fluid completes the composition of the whole mouth saliva with epithelial cells, leukocytes, serum transudate and multiple microorganisms. Through an active transport, extracellular ultrafiltration or passive diffusion, many blood constituents enter into the saliva, providing a new approach for the future development of a new diagnosis tool. Existing research shows the fact that certain circulating biomolecules that are specific to various diseases and were identified in the bloodstream, were as well present in the saliva of those patients [12]. This fact outlines the high functionality of using saliva as a diagnosis method, having the power to reflect different pathological states. Any alterations in the blood composition can lead to changes in the salivary biochemical composition.
The functions of saliva are based on its composition and the presents of its viscoelastic properties, and are presented by taste and smell, bolus formation, digestion, lubrication, wound healing, homeostasis, mineralization and buffer activity (Figure 2) [13].
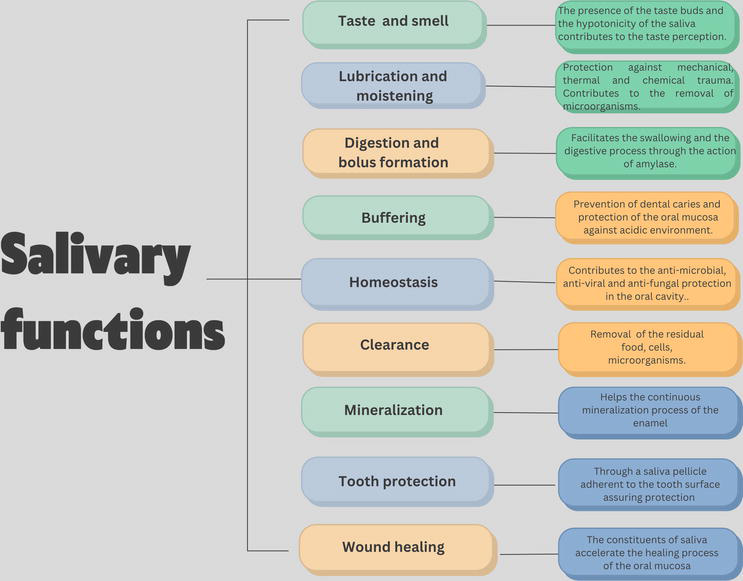
Figure 2.
Salivary functions and their description.
The human saliva offers an important source of proteins and peptides, that during the years proved their contribution in the diagnosis field, being association with diverse diseases. The recent studies reveled the implication of over one hundred biomolecules that could be identified in the salivary samples and fulfill the role of biomarkers for certain pathologies, including oral malignancy, diabetes, periodontal disease, dental caries, systemic disorders and other cancer types (Figure 3) [14, 15].
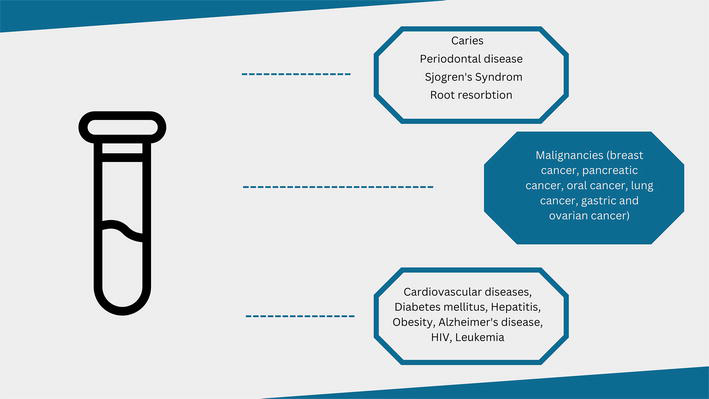
Figure 3.
Diagnostic use of saliva for local and general pathologies.
There are multiple options in collecting saliva, but the most recommended method is the collection of unstimulated saliva, several studies showing the fact that by using this type of collection method, the composition does not suffer any changes [16]. Nevertheless, regardless of the used approach to obtain a salivary sample, a clear protocol needs to be implemented regarding the storage, possible existent circadian variations and sample contamination in order to reflect correct information regarding the health of the individual [17]. The advantages provided by this type of sampling such as a non-invasive sampling and fast processing, easy storage and transportation, cost-effective, painless and easily accepted by patients make the use of saliva samples an option for the early diagnosis (Figure 4).
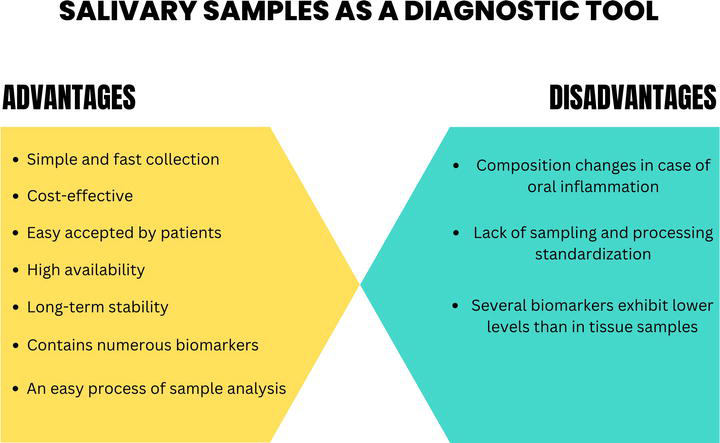
Figure 4.
Advantages and disadvantages of using salivary samples.
The notion of “Salivaomics” was introduced outlining the importance of analyzing the “omics” composition of saliva, focusing on the proteomic, metabolomic, genomic and transcriptomic particularities [18, 19]. A comprehensive analysis and identification of these components is an important part in the development of potential salivary biomarkers that could be linked to multiple pathologies [20]. As a diagnosis tool, saliva is a reliable biofluid that has the potential to replace the blood sample analysis, targeting the screening, early diagnosis and prognosis evaluation of multiple diseases, especially oral cancer [21].
3. Saliva as a diagnostic tool in oral cancer
Oral cancer, despite the advancements made in the diagnosis and therapeutic field, exhibits a continuous increase of its incidence, being mostly diagnosed in stages 3 or 4 accompanied by a poor prognosis [22]. An early detection of the oral potentially malignant disorders (OPMD) is important for a further prevention of oral malignancy development, improving the survival rate of the patients [23, 24, 25]. In order to avoid the diagnosis delay and invasiveness in case of performing a scalpel biopsy, the focus was directed toward developing non-invasive approaches [23, 26].
Salivary diagnostic is a current challenge for the research field, with a continuous development in the technology area introducing the screening techniques, next-generation sequencing, mass spectroscopy, proteomic and genomic analysis that can target even small quantities of potential oral cancer biomarkers [27]. The association of new technologies with the salivary diagnosis opened a new path toward discovering the presence of new molecules that can be linked to different carcinogenesis stages of oral cancer, focusing on screening, diagnosis, prognosis evaluation and post-treatment monitoring [28]. Multiple oral cancer biomarkers have been reported to be present in the saliva of the patients, especially in oral squamous cell carcinoma patients due to the fact that there are also altered analytes present and derived cells from the tumor environment. Nevertheless, the “omics” science outlined the contribution of the human saliva in the diagnosis field, offering a high number of analytes (metabolites, proteins and nucleic acids) and their alterations as a connection to a possible oral malignancy [29]. During the past years, many laboratory methods aimed to analyze and isolate specific salivary components that could be linked to the presence and evolution of diseases (Table 1).
| Type of analytical method | Identified salivary components | Reference |
|---|---|---|
| Enzyme-Linked Immunosorbent Assay (ELISA) | Proteins Virus Peptides Metabolites | [30, 31, 32] |
| PCR-based | Nucleic acids Bacteria Virus | [33, 34] |
| Raman-spectroscopy | Multiple biological molecules | [35] |
| Conductive Polymer Spray Ionization Mass Spectrometry (CPSI-MS) | Presence of drugs Metabolites | [36] |
| Matrix-Assisted Laser Desorption/Ionization- Time-of- Flight mass spectrometry (MALDI-TOF MS) | Proteins Microorganisms Peptides | [37] |
| High-Performance Liquid Chromatography (HPLC) coupled to Mass Spectrometry (MS) | Peptides Metabolites Proteins Hormones Vitamins | [38, 39, 40, 41] |
| Two-dimensional gel electrophoresis (2DE) coupled to mass spectrometry (MS) | Proteins | [42] |
| Electric Field-Induced Release and Measurement (EFIRM) | RNA Circulating single-stranded DNA | [43] |
| Nuclear Magnetic Resonance (1 H-NMR) spectroscopy | Metabolites | [44] |
| Attenuated Total Reflection-Fourier Transform Infrared (ATR-FTIR) spectroscopy | Nucleic acids Proteins Lipids | [45] |
Table 1.
3.1 Salivary proteomics in oral cancer diagnosis
Proteomics focuses on the study of the proteins that are expressed in the cells and tissue, providing important information related to a pathogenesis, with the possibility of being quantified through non-invasive samples, such as saliva. In case of oral cancer, the analysis of the proteins reveals important aspects regarding their functions, offering advantages over the tissue samples. The altered proteins identified in the OSCC carcinogenesis process are responsible for modifications in the cell structure, metabolism, cell motility and adhesion and intercellular signaling.
Multiple studies conducted complex analysis targeting the salivary proteome and its potential involvement in the diagnosis field of oral malignancy. The Salivary Proteome Knowledge Base and the information available in NIDCR’S Human Saliva Proteome contain valuable proteomic information provided also by the dataset resulted from the study conducted by Denny et al. [46]. The study conducted by Bandhakavi et al. [47] highlighted the existence of 2.340 proteins with direct involvement in different biological actions of the oral environment. An analysis was performed in order to compare the distribution of saliva and plasma proteins and concluded the fact that the analysis of salivary proteins has more advantages compared to the plasma proteins [48]. The identified salivary proteins that can be linked to oral cancer are mostly represented by interleukins, tumor-necrosis factor, CD44, CD59, IgG, p53 antibodies, transferrin, albumin, telomerase, alpha-amylase and S100 calcium-binding protein [49].
Along the past years, researcher’s attention was directed toward the existence of numerous proteins present in saliva that could be used for the early diagnosis of oral cancer, as well for evaluating the tumor evolution [50]. Studies focused on the implication of the interleukins (IL-6, IL-8, IL-1β), the tumor-necrosis factor alpha (TNF-α), transferrin, matrix metalloproteinase (MMP9, MMP2) and α-amylase [14].
Interleukins (IL) are a family of proteins that are responsible for the control of cellular migration, differentiation and the apoptosis process [51]. The pro-inflammatory category of cytokines IL-6, IL-8, IL-1 and TNF-α were identified in high levels in the salivary samples of OSCC patients and in case of premalignant lesions, outlining their potential role in the progression of the carcinogenesis process from a premalignant state to a malignant one [52]. Many studies have focused on the salivary levels of IL-8 that were encountered to be increased in periodontitis patients, but with a much higher level in case of oral cancer patients [53]. IL-8 and its high levels were associated with the tumor angiogenesis process, interfering in the cell circle and adhesion process [54]. Also, the salivary levels of IL-6 and TNF-α were reported to be increased in case of the presence of oral leukoplakia [27]. Nevertheless, Goldoni et al. [55] concluded that IL-8, IL-6, IL-1β and IL-1α and their salivary levels could represent a potential diagnosis tool for oral cancer. Research showed that a high number of proteins were identified to have increased levels in different stages of OSCC, being more relevant to focus on a specific one for a higher sensitivity [56]. Jou et al. [57] in their study revealed increased levels of transferrin in the saliva of OSCC patients, positively correlating them with the size and the stage of the tumor.
Recent studies showed that gelatinase-B (MMP-9) has an important diagnosis validity and had the highest sensitivity and specificity as a diagnosis tool for OSCC [58]. The cytokeratin fraction 21-1 (CYFRA 21-1) expression was positively correlated with OSCC, and studies revealed an important discriminating power between the potentially malignant disorder cases and OSCC [59].
3.2 Salivary metabolomics in oral cancer diagnosis
The pathogenesis of oral cancer is not completely understood, and studies discuss the importance of metabolic reprogramming as a future approach in the early diagnosis. During the tumorigenesis process, the cancer cells suffer important metabolic changes among which a metabolic reprogramming of the lipids and amino acids occurs [60].
The use of salivary metabolites has gained recent interest due to the fact that their presence in the saliva can be a consequence of the direct transfer from the tumor cells. Saliva metabolomics were used for assessing the early diagnosis and further monitorization of OSCC patients [61]. The analysis of the metabolites encountered in the saliva of the OSCC patients revealed increased levels of betaine, choline and pipecolic acid that were able to distinguish the OSCC patients from the control group [62].
A study performed by Sugimoto et al. [63] in which they analyzed the saliva collected from oral cancer patients identified 57 metabolites that could be linked to this malignancy. When compared to the control group, the salivary levels of polyamines were significantly higher [63], other studies discussing the fact that the increased levels determined a high cell proliferation rate, promote tumor invasiveness and decrease the apoptosis process [64]. Also, the presence of piperidine and taurine showed a high potential for their use as oral screening tool [62]. Another study that focused on the salivary metabolome as a diagnosis tool outlined the role of putrescine for monitoring the action of chemotherapy in oral cancer [65].
Wei et al. [66] in their study aimed to evaluate the salivary metabolomics and their changes in OSCC, leukoplakia and oral lichen planus patients and identified five metabolites with the power to discriminate between the three groups of patients, among which were lactate, valine, phenylalanine, n-eicosanoic acid and γ-aminobutyric acid.
In order to establish and certify the presence of the altered salivary metabolites and their correlation with oral cancer, Ishikawa et al. [67] conducted a study in which they compared the altered metabolites encountered in tissue and salivary samples of OSCC patients. Their results showed that 17 metabolites had altered levels in both types of samples, and pipecolate and
Nevertheless, the diagnosis potential of salivary metabolites is still a discussed topic, as there are certain differences in the salivary metabolome dependent on gender, type of saliva (stimulated or unstimulated), associated risk factors (such as smoking), circadian variation of the salivary composition and age [68]. The stability of these metabolites is still questionable and in order to accomplish their role as a diagnostic tool, a proper protocol needs to be implemented that should limit the possible variations related to them.
3.3 Salivary genomics in oral cancer diagnosis
The presence of cell-free circulation DNA (deoxyribonucleic acid) in the plasma was outlined approximately 60 years ago [69], and its altered levels were identified in cancer patients compared to healthy controls, exhibiting specific tumoral characteristics [70]. These alterations are represented by abnormal methylation, somatic mutations in oncogenes and tumor suppressor genes, mitochondrial DNA mutations, microsatellite alterations and viral tumor-related DNA [71]. Studies have shown that the body fluids, saliva included, contain cell-free nucleic acids with an important role in the diagnosis field. Saliva-based tests have reflected a high potential in detecting oral cancer by analyzing the hypermethylation levels, oral microbiota and existent exfoliated cells [69].
In the human saliva, the total DNA content was reported to be between 1.8 and 128 μg/mL, and approximately, 70% is from the host and 30% has its origins from the oral microbiome [72]. Research has outlined the fact that the quality of the salivary DNA was from 72% up to 96% in the samples that were genotyped [73].
Multiple studies focused on revealing specific genetic alteration that could be associated with the development, progression and metastasis rate of oral cancer. There has been reported the existence of a tumor-specific genome, among which the alterations of p53 and the presence of tumor suppressor gene mutations were directly linked to the oral malignancy carcinogenesis process [74]. A loss of heterozygosity (LOH) was revealed, representing a loss of genomic material localized on one of the chromosomes. Based on this fact, studies showed that specific LOH in regions that are responsible tumor suppressor genes is associated and represents an indicator for the malignant transformation in case of oral premalignant lesions [75]. It was reported that frequent LOH was identified in chromosomes 3q, 9p, 13q and 17p was linked to an early onset of oral malignancy [76]. Mutations that occurred in the mitochondrial DNA showed their implication in identifying exfoliated OSCC cells in salivary samples [77]. The hypermethylation of several genes was discovered and linked to the occurrence of cancers in the head and neck area. The study conducted by Rosas et al. [78] showed abnormal methylation encountered in certain genes (DAP-K, MGMT and p16) in patients diagnosed with OSCC, and outlined their potential for the detection and further recurrence surveillance of the oral malignancy. Liao et al. [79] identified the mutation of p53 gene in the saliva of OSCC patients, with a high potential role in the early detection of OSCC. A study conducted by Zhong et al. [80] aimed to detect the telomerase activity in the saliva of OSCC diagnosed patients and its potential as a marker.
Existent research outlines the fact that certain genomic alterations that were identified in the tumor tissue samples were also present in the salivary samples. Mutations in CDKN2A, PIKC3, TP53 and NOTCH1, along with translocation mutations in tumor DNA were discovered in the salivary samples of OSCC patients [81]. These results highlight the idea that higher levels of remanent tumor DNA are encountered in saliva samples compared to plasma ones, with a high contribution in the further development of non-invasive personalized early diagnosis.
3.4 Salivary transcriptomics in oral cancer diagnosis
Transcriptomics represents the analysis of a complex set of RNA (ribonucleic acid) transcripts that are present in certain circumstances and are discovered through technologies such as polymerase chain reaction (PCR), microarray technology and next-generation sequencing. In cell-free whole saliva, the total RNA level ranged between from 0.108 ± 0.023 μg/mL [82]. It was identified that the majority of the cell-free RNAs from whole mouth saliva are human-derived mRNAs [83]. Approximately 3000 to 6400 human mRNAs were detectable in saliva and 27.5% of the investigated mRNAs have still unknown functions [84]. Messenger RNA represents a direct precursor of proteins, and its levels usually are corresponding and can be correlated. The nucleic acids are considered easier to screen and offer a viable option to become a disease marker.
The dysregulation of different miRNAs controls the cell differentiation process, growth and apoptosis, influencing tumor suppressors and oncogenes [54]. Salazar et al. [85] in their study identified elevated levels of salivary miR-134, miR-9 and miR-191 in the samples of oral cancer patients, suggesting their use as potential markers. Other study reported decreased levels of miR-200a and miR-125a [86] and high levels of miR-31 in the saliva of diagnosed OSCC patients. miR-31 was linked to the post-treatment monitoring of OSCC patients, as results reveal the fact that the salivary levels decreased after the tumor resection [86]. Another study performed by Li et al. [87] by using the microarray analysis revealed significantly modified levels in seven potential markers for oral cancer, represented by the increased regulation of three groups of mRNAs (IL-8, S100P and H3F3A), while decreased mRNA levels were noted in SAT, OAZ1 and DUSP1.
A concern was the stability of salivary miRNAs and the protocol related to the collection and time of processing the samples in order to provide an accurate molecular profiling analysis. Zimmermann et al. [69] in their study focused on evaluating the salivary cell-free miRNA and their finding outline the existence of a salivary core transcriptome of 185 gene that were identified in all the included samples.
4. Conclusions
Taking into consideration the current epidemiological trend that describes an increase of oral cancer incidence globally, the prevention and early diagnosis strategies need to overcome the existing limits and disadvantages that are influencing the survival rate of these patients.
Saliva represents a promising tool for the early diagnosis of oral cancer, providing also important information related to the progression and post-treatment surveillance. The advancements made in technology in order to assure a complex analysis of the salivary samples allowed researchers to expand the use of saliva as a diagnosis tool and validate multiple components with direct implications in the oral carcinogenesis process. The diagnosis field urges the need to implement non-invasive approaches for screening and early diagnosis, and currently, saliva offers multiple advantages such as a non-invasiveness, easy sampling, painless and fast collection.
By implementing the” omics” concept, saliva testing becomes a reliable and appealing body fluid. Studies have shown that especially related to oral cancer, specific components that exist or are delivered by the tumor, can act like disease markers and are present in a higher percentage in saliva. Current progress has been made in order to quantify and understand their characteristics. An important aspect that should be mentioned is the need to implement a protocol that stands for a standardization of the salivary collection method, storage and analysis, minimizing the possible existent factors that could intervene in becoming a proper diagnosis tool.
The oral carcinogenesis process is a complex one, with specific alterations at every evolutive stage, salivary composition providing in many cases an individual perspective on the ongoing changes, having a direct contact with the tumoral environment. Despite the existing limitations, the salivary diagnostics promise an increase in the clinical applicability, targeting specific proteomic, genomic and transcriptomic alterations that are associated with the oral malignancy.
References
- 1.
Madhura MG, Rao RS, Patil S, Fageeh HN, Alhazmi A, Awan KH. Advanced diagnostic aids for oral cancer. Disease-a-Month. 2020; 66 :1-12 - 2.
Neville BW, Day TA. Oral cancer and precancerous lesions. CA: A Cancer Journal for Clinicians. 2002; 52 :195-215 - 3.
Wong DT. Towards a simple, saliva-based test for the detection of oral cancer. ‘Oral fluid (saliva), which is the mirror of the body, is a perfect medium to be explored for health and disease surveillance’. Expert Review of Molecular Diagnostics. 2006; 6 :267-272 - 4.
Manoharan S, Karthikeyan S, Essa MM, Manimaran A, Selvasundram R. An overview of oral carcinogenesis. International Journal of Nutrition, Pharmacology, Neurological Diseases. 2016; 6 (2):51-62 - 5.
Nambiar KS, Haragannavar VC, Augustine D, Sowmya SV, Rao RS. Diagnostic aids in detection of oral precancer and cancer: Past to present. International Dental & Medical Journal of Advanced Research. 2016; 2 :1-7 - 6.
Chaudhary R, Shah A, Shah DM, Singh S, Thakkar P, Goyal S. Advanced diagnostic aids in detection of oral cancer. International Journal of Dental and Medical Research. 2014; 1 (3):139-143 - 7.
Dawes C. Rhythms in salivary flow rate and composition. International Journal of Chronobiology. 1974; 2 :253-279 - 8.
Mese H, Matsuo R. Salivary secretion, taste and hyposalivation. Journal of Oral Rehabilitation. 2007; 34 :711-723 - 9.
Walsh NP, Montague JC, Callow N, Rowlands AV. Saliva flow rate, total protein concentration and osmolality as potential markers of whole body hydration status during progressive acute dehydration in humans. Archives of Oral Biology. 2004; 49 :149-154 - 10.
Chiappin S, Antonelli G, Gatti R, De Palo EF. Saliva specimen: A new laboratory tool for diagnostic and basic investigation. Clinica Chimica Acta. 2007; 383 :30-40 - 11.
Kaczor-Urbanowicz KE, Wei F, Rao SL, Kim J, Shin H, Cheng J, et al. Clinical validity of saliva and novel technology for cancer detection. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 2019; 1872 (1):49-59 - 12.
Pfaffe T, Cooper-White J, Beyerlein P, Kostner K, Punyadeera C. Diagnostic potential of saliva: Current state and future applications. Clinical Chemistry. 2011; 57 :675-687 - 13.
Carpenter GH. The secretion, components, and properties of saliva. Annual Review of Food Science and Technology. 2013; 4 :267-276 - 14.
Zhang CZ, Cheng XQ , Li JY, Zhang P, Yi P, Xu X, et al. Saliva in the diagnosis of diseases. International Journal of Oral Science. 2016; 8 (3):133-137 - 15.
Sultan AS, Kong EF, Jabra-Rizk MA. The oral microbiome: A lesson in co-existence. PLoS Pathogens. 2018; 14 (1):e1006719 - 16.
Kaczor-Urbanowicz KE, Martin Carreras-Presas C, Aro K, Tu M, Garcia-Godoy F, Wong DT. Saliva diagnostics - current views and directions. Experimental Biology and Medicine (Maywood). 2017; 242 (5):459-472. DOI: 10.1177/1535370216681550 - 17.
Kaur J, Srivastava R, Borse V. Recent advances in point-of-care diagnostics for oral cancer. Biosensors and Bioelectronics. 2021; 178 :112995 - 18.
Wong DT. Salivaomics. Journal of the American Dental Association (1939). 2012; 143 (Suppl. 10):19S-24S - 19.
Tasoulas J, Patsouris E, Giaginis C, Theocharis S. Salivaomics for oral diseases biomarkers detection. Expert Review of Molecular Diagnostics. 2016; 16 (3):285-295 - 20.
Wang X, Kaczor-Urbanowicz KE, Wong DT. Salivary biomarkers in cancer detection. Medical Oncology. 2017; 34 :7 - 21.
Roblegg E, Coughran A, Sirjani D. Saliva: An all-rounder of our body. European Journal of Pharmaceutics and Biopharmaceutics. 2019; 142 :133-141 - 22.
Wu DT, Tao O, Trinh N, Javaid MA, Ahmed AS, Durand R, et al. Saliva–a promising tool for diagnosing oral diseases. Current Oral Health Reports. 2018; 5 (4):242-249 - 23.
Liu D, Zhao X, Zeng X, Dan H, Chen Q. Non-invasive techniques for detection and diagnosis of oral potentially malignant disorders. The Tohoku Journal of Experimental Medicine. 2016; 238 (2):165-177 - 24.
Walsh T, Macey R, Brocklehurst P, Kerr AR, Liu JL, Lingen MW, et al. Diagnostic tests for oral cancer and potentially malignant disorders in patients presenting with clinically evident lesions. Cochrane Database of Systematic Reviews. 2015; 5 :CD010276 - 25.
Stuani VT, Rubira CM, Sant’Ana AC, Santos PS. Salivary biomarkers as tools for oral squamous cell carcinoma diagnosis: A systematic review. Head & Neck. 2017; 39 (4):797-811 - 26.
Panta P, Venna VR. Salivary RNA signatures in oral cancer detection. Analytical Cellular Pathology. 2014; 2014 :7-7 - 27.
Goldoni R, Scolaro A, Boccalari E, Dolci C, Scarano A, Inchingolo F, et al. Malignancies and biosensors: A focus on oral cancer detection through salivary biomarkers. Biosensors. 2021; 11 :396 - 28.
Khurshid Z, Zafar MS, Khan RS, Najeeb S, Slowey PD, Rehman IU. Role of salivary biomarkers in oral cancer detection. Advances in Clinical Chemistry. 2018; 86 :23-70 - 29.
Thomadaki K, Helmerhorst EJ, Tian N, Sun X, Siqueira WL, Walt DR, et al. Whole-saliva proteolysis and its impact on salivary diagnostics. Journal of Dental Research. 2011; 90 :1325-1330 - 30.
Desrosiers NA, Huestis MA. Oral fluid drug testing: Analytical approaches, issues and interpretation of results. Journal of Analytical Toxicology. 2019; 43 :415-443 - 31.
Messana I, Cabras T, Iavarone F, Vincenzoni F, Urbani A, Castagnola M. Unraveling the different proteomic platforms. Journal of Separation Science. 2013; 36 :128-139 - 32.
Chu HW, Chang KP, Hsu CW, Chang IY, Liu HP, Chen YT, et al. Identification of salivary biomarkers for oral cancer detection with untargeted and targeted quantitative proteomics approaches. Molecular & Cellular Proteomics. 2019; 18 :1796-1806 - 33.
Jamieson LM, Antonsson A, Garvey G, Ju X, Smith M, Logan RM, et al. Prevalence of oral human papillomavirus infection among Australian indigenous adults. JAMA Network Open. 2020; 3 :e204951 - 34.
Fadhil RS, Wei MQ , Nikolarakos D, Good D, Nair RG. Salivary microRNA miR-let-7a-5p and miR-3928 could be used as potential diagnostic bio-markers for head and neck squamous cell carcinoma. PLoS One. 2020; 15 :e0221779 - 35.
Lin D, Yang SW, Hsieh CL, Hsu KJ, Gong T, Wu Q , et al. Tandem quantification of multiple carbohydrates in saliva using surface-enhanced Raman spectroscopy. ACS Sensors. 2021; 6 :1240-1247 - 36.
Song X, Yang X, Narayanan R, Shankar V, Ethiraj S, Wang X, et al. Oral squamous cell carcinoma diagnosed from saliva metabolic profiling. Proceedings of the National Academy of Sciences of the United States of America. 2020; 117 :16167-16173 - 37.
Antezack A, Chaudet H, Tissot-Dupont H, Brouqui P, Monnet-Corti V. Rapid diagnosis of periodontitis, a feasi- bility study using MALDI-TOF mass spectrometry. PLoS One. 2020; 15 :e0230334 - 38.
Alvi SN, Hammami MM. An improved method for measurement of testosterone in human plasma and saliva by ultra-performance liquid chromatography-tandem mass spectrometry. Journal of Advanced Pharmaceutical Technology & Research. 2020; 11 :64-68 - 39.
Wang Q , Yu Q , Lin Q , Duan Y. Emerging salivary biomarkers by mass spectrometry. Clinica Chimica Acta. 2015; 438 :214-221 - 40.
Wu CC, Chu HW, Hsu CW, Chang KP, Liu HP. Saliva proteome profiling reveals potential salivary biomarkers for detection of oral cavity squamous cell carcinoma. Proteomics. 2015; 15 :3394-3404 - 41.
Kawahara R, Bollinger JG, Rivera C, Ribeiro AC, Brandão TB, Paes Leme AF, et al. A targeted proteomic strategy for the measurement of oral cancer candidate biomarkers in human saliva. Proteomics. 2016; 16 :159-173 - 42.
Jessie K, Jayapalan JJ, Ong KC, Abdul Rahim ZH, Zain RM, Wong KT, et al. Aberrant proteins in the saliva of patients with oral squamous cell carcinoma. Electrophoresis. 2013; 34 :2495-2502 - 43.
Cheng J, Nonaka T, Wong DTW. Salivary exosomes as nanocarriers for cancer biomarker delivery. Materials. 2019; 12 :654 - 44.
Herrala M, Mikkonen JJW, Pesonen P, Lappalainen R, Tjäder- Niemelä RK, Seitsalo H, et al. Variability of salivary metabolite levels in patients with Sjögren’s syndrome. Journal of Oral Science. 2020; 63 :22-26 - 45.
Ferreira IC, Aguiar EM, Silva AT, Santos LL, Cardoso-Sousa L, Araújo TG, et al. Attenuated total reflection-Fourier transform infrared (ATR-FTIR) spectroscopy analysis of saliva for breast cancer diagnosis. Journal of Oncology. 2020; 2020 :4343590 - 46.
Denny P, Hagen FK, Hardt M, Liao L, Yan W, Arellanno M, et al. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. Journal of Proteome Research. 2008; 7 (5):1994-2006 - 47.
Bandhakavi S, Stone MD, Onsongo G, Van Riper SK, Griffin TJ. A dynamic range compression and three-dimensional peptide fractionation analysis platform expands proteome coverage and the diagnostic potential of whole saliva. Journal of Proteome Research. 2009; 8 (12):5590-5600 - 48.
Loo JA, Yan W, Ramachandran P, Wong DT. Comparative human salivary and plasma proteomes. Journal of Dental Research. 2010; 89 (10):1016-1023 - 49.
Khurshid Z, Zohaib S, Najeeb S, Zafar M, Rehman R, Rehman I. Advances of proteomic sciences in dentistry. International Journal of Molecular Sciences. 2016; 17 :728 - 50.
Chundru VNS, Nirmal RM, Srikanth B, Bojji M, Midhun N, Lakshmi BJ. Salivaomics for oral cancer detection: An insight. Journal of Pharmacy & Bioallied Sciences. 2021; 13 :S52-S56 - 51.
Tasoulas J, Patsouris E, Giaginis C, Theocharis S. Salivaomics for oral diseases biomarkers detection. Expert Review of Molecular Diagnostics. 2016; 16 :285-295 - 52.
Papale F, Santonocito S, Polizzi A, Giudice AL, Capodiferro S, Favia G, et al. The new era of salivaomics in dentistry: Frontiers and facts in the early diagnosis and prevention of oral diseases and cancer. Metabolites. 2022; 12 :638 - 53.
Buzalaf MAR, Ortiz AC, Carvalho TS, Fideles SOM, Araújo TT, Moraes SM, et al. Saliva as a diagnostic tool for dental caries, periodontal disease and cancer: Is there a need for more biomarkers? Expert Review of Molecular Diagnostics. 2020; 20 :543-555 - 54.
Radhika T, Jeddy N, Nithya S, Muthumeenakshi RM. Salivary biomarkers in oral squamous cell carcinoma—An insight. Journal of Oral Biology and Craniofacial Research. 2016; 6 :S51-S54 - 55.
Govindraju P, Kumar T. Genomic alphabets of saliva as a biomarker in oral cancer. Journal of Indian Academy of Oral Medicine and Radiology. 2017; 29 :300 - 56.
Elashoff D, Zhou H, Reiss JK, Wang J, Henson B, Hu S, et al. Pre-validation of salivary biomarkers for oral cancer detection. Cancer Epidemiology, Biomarkers & Prevention. 2012; 21 (4):664-672 - 57.
Jou YL, Lin CD, Lai CH, Chen CH, Kao JY, Chen SY, et al. Proteomic identification of salivary transferrin as a biomarker for early detection of oral cancer. Analytica Chimica Acta. 2010; 681 :41-48 - 58.
Ghallab NA, Shaker OG. Serum and salivary levels of chemerin and MMP-9 in oral squamous cell carcinoma and oral premalignant lesions. Clinical Oral Investigations. 2017; 21 :937-947 - 59.
Rajkumar K, Ramya R, Nandhini G, Rajashree P, Ramesh Kumar A, Nirmala AS. Salivary and serum level of CYFRA 21-1 in oral cancer and oral squamous cell carcinoma. Oral Diseases. 2015; 21 :90-96 - 60.
Yang LF, Venneti S, Nagrath D. Glutaminolysis: A hallmark of cancer metabolism. Annual Review of Biomedical Engineering. 2017; 19 :163-194 - 61.
Mikkonen JJ, Singh SP, Herrala M, Lappalainen R, Myllymaa S, Kullaa AM. Salivary metabolomics in the diagnosis of oral cancer and periodontal diseases. Journal of Periodontal Research. 2016; 2016 (51):431-437 - 62.
Wang Q , Gao P, Wang X, Duan Y. Investigation and identification of potential biomarkers in human saliva for the early diagnosis of oral squamous cell carcinoma. Clinica Chimica Acta. 2014; 427 :79-85 - 63.
Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010; 6 :78-95 - 64.
Gerner EW, Meyskens FL. Polyamines and cancer: Old molecules, new understanding. Nature Reviews Cancer. 2004; 4 :781-792 - 65.
Okamura M, Kobayashi M, Suzuki F, Shimada J, Sakagami H. Induction of cell death by combination treatment with cisplatin and 5-fluorouracil in a human oral squamous cell carcinoma cell line. Anticancer Research. 2007; 27 :3331-3337 - 66.
Wei J, Xie G, Zhou Z, Shi P, Qui Y, Zheng X, et al. Salivary metabolite signatures of oral cancer and leukoplakia. International Journal of Cancer. 2011; 129 :2207-2217 - 67.
Ishikawa S, Sugimoto M, Kitabakate K, Tu M, Sugano A, Yamamori I, et al. Effect of timing of collection of salivary metabolomic biomarkers on oral cancer detection. Amino Acids. 2017; 49 :761-770 - 68.
Takeda I, Stretch C, Barnaby P, Bhatnager K, Rankin K, Fu H, et al. Understanding the human salivary metabolome. NMR in Biomedicine. 2009; 22 :577-584 - 69.
Zimmermann BG, Park NJ, Wong DT. Genomic targets in saliva. Annals of the New York Academy of Sciences. 2007; 1098 (1):184-191 - 70.
Leon SA, Shapiro B, Sklaroff DM, Yaros. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Research. 1977; 37 :646-650 - 71.
Anker P, Mulcahy H, Chen XQ , Stroun M. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metastasis Reviews. 1999; 18 :65-73 - 72.
Hansen TO, Simonsen MK, Nielsen FC, Hundrup YA. Collection of blood, saliva, and buccal cell samples in a pilot study on the Danish nurse cohort: Comparison of the response rate and quality of genomic sample. Cancer Epidemiology, Biomarkers & Prevention. 2007; 16 :2072-2076 - 73.
Rylander-Rudquist T, Hakansson N, Tybring G, Wolk A. Quality and quantity of saliva DNA obtained from the self-administered oragene method - A pilot study on the cohort of Swedish men. Cancer Epidemiology, Biomarkers & Prevention. 2006; 15 :1742-1745 - 74.
Shah FD, Begum R, Vajaria BN, Patel KR, Patel JB, Shukla SN, et al. A review on salivary genomics and proteomics biomarkers in oral cancer. Indian Journal of Clinical Biochemistry. 2011; 26 :326-334 - 75.
Fliss MS, Usadel H, Caballero OL, Wu L, Buta MR, Eleff SM, et al. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000; 287 :2017-2019 - 76.
Zhang L, Rosin MP. Loss of heterozygosity: A potential tool in management of oral premalignant lesions? Journal of Oral Pathology & Medicine. 2001; 30 :513-520 - 77.
Jiang WW, Masayesva B, Zahurak M, Carvalho AL, Rosenbaum E, Mambo E, et al. Increased mitochondrial DNA content in saliva associated with head and neck cancer. Clinical Cancer Research. 2005; 11 :2486-2491 - 78.
Rosas SLB, Koch W, Da Costa Carvalho MDG, Wu L, Califano J, Westra W, et al. Promoter hypermethylation patterns of p16, O6-methylguanine- DNA-methyltransferase, and death-associated protein kinase in tumors and saliva of head and neck cancer patients. Cancer Research. 2001; 61 :939-942 - 79.
Liao PH, Chang YC, Huang MF, Tai KW, Chou MY. Mutation of p53 gene codon 63 in saliva as a molecular marker for oral squamous cell carcinomas. Oral Oncology. 2000; 36 :272-276 - 80.
Zhong LP, Chen GF, Xu ZF, Zhang X, Ping FY, Zhao SF. Detection of telomerase activity in saliva from oral squamous cell carcinoma patients. International Journal of Oral and Maxillofacial Surgery. 2005; 34 :566-570 - 81.
Zandberg DP, Tallon LJ, Nagaraj S, Sadzewicz LK, Zhang Y, Strome MB, et al. Intratumor genetic heterogeneity in squamous cell carcinoma of the oral cavity. Head & Neck. 2019; 41 :2514-2524 - 82.
Feng L, Houck JR, Lohavanichbutr P, Chen C. Transcriptome analysis reveals differentially expressed lncRNAs between oral squamous cell carcinoma and healthy oral mucosa. Oncotarget. 2017; 8 :31521-31531 - 83.
Li Y, Zhou X, St. John MAR, Wong DTW. RNA profiling of cell-free saliva using microarray technology. Journal of Dental Research. 2004; 83 :199-203 - 84.
Park NJ, Zhou X, Yu T, Brinkman BMN, Zimmermann BG, Palanisamy V, et al. Characterization of salivary RNA by cDNA library analysis. Archives of Oral Biology. 2007; 52 :30-35 - 85.
Salazar C, Nagadia R, Pandit P, Cooper-White J, Banerjee N, Dimitrova N, et al. A novel saliva-based microRNA biomarker panel to detect head and neck cancers. Cellular Oncology. 2014; 37 :331-338 - 86.
Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, Abemayor E, et al. Salivary microRNA: Discovery, characterization, and clinical utility for oral cancer detection. Clinical Cancer Research. 2009; 15 :5473-5477 - 87.
Li Y, St. John MA, Zhou X, Kim Y, Sinha U, Jordan RC, et al. Salivary transcriptome diagnostics for oral cancer detection. Clinical Cancer Research. 2004; 10 :8442-8450