Breakdown of patients A-D RIN values [18].
Abstract
Rapid postmortem tissue collection has been shown to have increasing use for molecular and genetic profiling. Although research on human tissue has been conducted for many years, modern molecular assays have significantly higher sensitivity and specificity than those used in the past. Higher grade tissue specimens are now required for the extraction of macromolecules as a result of this. In fact, these studies have paved the way for multiple postmortem tissue collection studies such as COVID-19, brain, skin, and small cell lung cancer. Best practices for tissue collection have also been developed by a number of professional organizations, including the National Institutes of Health Office of Biorepositories and the International Society of Biological and Environmental Repositories (ISBER). These guidelines can be used to establish procedures for tissue collection.
Keywords
- autopsy
- post-mortem
- COVID-19
- small cell lung cancer
- SCLC
- skin
- histology
- post-mortem interval
- RIN
1. Introduction
One of the most important tools for the study of human disease and the creation of new therapeutics is human tissue specimens. The techniques utilized for sample collecting and processing can have a significant impact on the molecular information included in tissue samples. Several factors that are essential for collecting high-quality tissues are covered in this chapter. By adhering to pre-established guidelines and standard operating procedures, tissue samples will be handled consistently and optimally, minimizing the impact of environmental factors on a sample’s quality and molecular profile.
2. Tissue collection for genomic research
2.1 Tissue collection
Proper collection and preservation of tissue specimens is fundamental to the ability to isolate enough high-quality nucleic acids for genomic research. It is now well-recognized that changes in the molecular profile of a tissue can occur during sample retrieval, between the time interval from retrieval to processing, and during processing itself. Therefore, it is important to establish standardized guidelines that keep the time from specimen collection to preservation as short as possible and maintain consistency in the handling of samples.
One of the major factors influencing the integrity of nucleic acids in stored tissue samples is tissue ischemia time. The surgical disruption of blood flow to an excised clinical sample leads to progressive tissue ischemia, hypoxia, and release of endogenous nuclease enzymes capable of degrading nucleic acids. The warm ischemia time is the time from the interruption of the blood supply to the removal of a tissue sample from the patient, while the cold ischemia time is the time from removal of a tissue to its preservation by freezing or by placement in a fixative. Warm ischemia time is dependent on the surgery and can be difficult to control. In contrast, the institution’s tissue collection and transportation protocols affect the cold ischemia time and can be readily controlled using standardized sample collection and handling procedures. To minimize tissue and cell damage, the cold ischemia time should be kept as short as possible. It is important that both the warm and cold ischemia times are documented [1, 2].
2.2 Tissue preservation and storage
In general, three basic methods of sample preservation exist: snap freezing, fixation, and a nucleic acid stabilizing reagent. If enough material is available, it is often advisable to split and preserve the tissue sample using multiple methods (e.g. snap freeze half the tissue and fix the other half in formalin).
Fresh-frozen tissue samples provide optimal quality nucleic acids, compared to fixed tissue specimens. However, the freezing process can compromise tissue morphology, so this method is not always ideal if tissue morphology is to be examined. The process of snap-freezing may also pose logistical issues for some laboratories because it requires specialized equipment. Snap-freezing (also known as flash freezing) is the process by which samples are lowered to temperatures below −70°C very rapidly. Snap-freezing, as opposed to standard freezing, reduces the chances of water in the sample forming ice crystals and damaging the tissue. Snap-freezing also slows the actions of nucleases and proteases thus inhibiting the degradation of nucleic acids and protein in the sample.
If examination of tissue morphology by histochemical or immunohistochemical means is required, formalin fixation and paraffin embedding (FFPE) is the standard process for sample preservation. The FFPE process not only preserves tissue morphology, but also allows for easy transport and storage and permanent preservation of the tissue. Formalin fixation is carried out by submerging the tissue specimen directly in 10% neutral-buffered formalin (NBF) at room temperature. For molecular assays, the optimal fixation time is approximately 14–18 h. Studies have shown that longer fixation times tend to result in poor quality RNA.
Another common method of preservation is submersion of the tissue directly in a nucleic acid stabilizing reagent such as PAXgene® Tissue Fix, TRIzol or RNAlater. This method is ideal for minimizing nucleic acid damage and changes in gene expression.
2.3 Use of wet ice for stabilization of genomic RNA
One method of transporting organs is through the use of wet ice. One pilot cardiovascular study collecting explanted hearts from 1996 to 2008 experimented using this method. There were 50 heart explants from the heart transplant team. The explanted hearts were delivered to the surgical pathology gross room by the heart team. The heart was then packed in a stainless-steel surgical bucket of wet crushed ice using the three-bag technique. The heart was washed in transplant solution before putting the first bag in preservation solution which was patented by University of Wisconsin. The heart was dissected with approximately half the sections being transferred into 10% NBF and the other half into small containers surrounded by wet ice for transportation to the processing lab. The tissue samples were placed in an −80° C freezer and put into a liquid nitrogen tank. Research at the time did not fully support the use of wet ice for preservation of RNA; however, this study found that the RIN quality was not impacted by using wet ice. Other studies worked with four different organ procurement organizations (OPO) in Indianapolis for about 10 years and found the same preservation of RNA in wet ice yielded high RIN values [3, 4, 5, 6, 7].
2.4 Histology quality control
Quality control analysis begins at the histology level and should be performed real-time as tissue specimens are received. For each procured tissue, a hematoxylin and eosin (H&E)-stained slide should be generated and reviewed under a microscope. In the case of tumor specimens, the pathologist confirms that the sample contains enough tumor cells (at least 50%) to be considered acceptable for molecular analysis. Tumor samples that contain less than 50% tumor might be released only for applications, such as immunohistochemistry or in situ hybridization. In addition to evaluating the percentage of viable tumor cells, guidelines issued by ISBER and NCI state the pathologist should also make note of the percentages of necrosis, fibrosis, mucin formation, stroma, normal adjacent tissue, and inflammation [8, 9, 10, 11, 12, 13].
2.5 Nucleic acid quality control
Archival FFPE samples are notorious for yielding poor-quality nucleic acids after 5 years of storage. Therefore, it is important to evaluate the molecular quality of each sample before its release for subsequent molecular testing. For retention of high-quality RNA, freezing is currently the best method of long-term tissue storage. Although not as labile as RNA, DNA molecules are also susceptible to degradation during the long-term storage of tissue samples [8, 9, 10].
2.6 Post-mortem specimens
Post-mortem human tissues are a valuable source of biological material. These specimens have versatile research applications, but the quality must be maintained throughout processing. In animal studies, the handling and storage of tissues immediately after death can be easily controlled. However, in human studies, the uncontrollability of these factors leads to a higher degree of variability. For example, the time lag between death and preservation of a sample following autopsy (PMI) is a major factor influencing the quality of RNA in post-mortem tissues. Existing evidence suggests that there is little correlation between the post-mortem interval and RNA quality/RIN score. The integrity of RNA in post-mortem samples appears to be linked to pre-mortem factors such as the cause of death and the length and duration of the period just before death, known as the agonal period.
3. Background of pilot RNA post-mortem study
3.1 Introduction
Numerous research studies have analyzed the post-mortem brain tissue’s quality. When analyzing post-mortem brain tissue, the goal of genetic research has been to comprehend neurological illnesses. These investigations have made significant progress and identified high RNA quality [14, 15]. These studies recorded pre-mortem fever or sepsis, pre-mortem tissue pH, temporal relationship to time of death (either sudden death or protracted illness), post-mortem interval, and other significant health conditions prior to death [16]. In the last 10 years, the integrity and general quality of the tissue have been assessed using the quality of the RNA. RNA quality is currently assessed using electrophoresis and RIN values [17]. The length of time before death till tissue removal, the classification of death, and the state of the patient’s health before death all may have an impact on the quality of the RNA in the tissue [18].
In addition to RNA quality assessed by RIN values, the area of postmortem neuropathology has developed chemical markers for tissue quality, starting with pH and the 28S/18S ratio. Ten organ tissues from four postmortem instances with various PMI were examined. To ascertain the histology and molecular markers of the postmortem tissue, the clinical history, agonal condition, refrigeration time, and PMI of 10, 12, 16, and >24 h were assessed (Figure 1) [18].
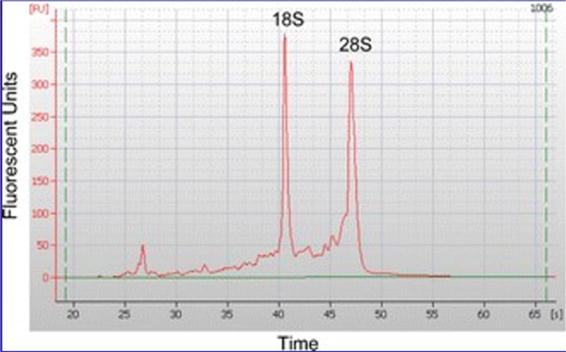
Figure 1.
Tissue RNA evaluation using a whole electrophoretogram to quantitate a RIN value.
3.2 Results
RNA stability (RIN values) was not predicted by the PMI, although cooling after death affected body temperature, postponed post-mortem autolysis, and improved RIN levels. However, RIN values fell with sustained hypoxemia. Two of the key factors influencing RIN values were agonal state scores and clinical medical history. RIN values decreased when agonal state scores were high, which is consistent with previous research. Most organs’ RIN scores could be predicted from histology, with the exception of the pancreas and small intestine, which autolyzed relatively quickly. RIN values for all organs are shown in Table 1.
| Tissue | A | B | C | D | Average |
|---|---|---|---|---|---|
| Liver | 3 | 4.9 | 2.5 | 7.7 | 4.5 |
| Kidney | 7.2 | 2.9 | 3.4 | 4.1 | 4.4 |
| Heart | 8.2 | 6.8 | 7.7 | 5.8 | 7.1 |
| Lung | 5.6 | 5.8 | 5.1 | 7 | 5.9 |
| Small Intestine | 5.2 | 6.4 | 3.6 | 5.5 | 5.2 |
| Pancreas | 4.7 | 2.6 | 1.8 | 2.7 | 3 |
| Skin | 7.4 | 3.6 | 4.6 | 7.8 | 5.9 |
| Brain | 7.9 | 4.5 | 4.7 | N/C | N/C |
| Cerebellum | 8.7 | 3.9 | N/C | 5.3 | N/C |
| Skeletal muscle | 8.3 | N/C | N/C | N/C | N/C |
| Bone marrow | 2.7 | 2.9 | N/C | 3 | N/C |
Table 1.
This study was used as a pilot for the following ongoing human genomic studies: suicidality [19], small cell lung cancer, COVID-19 [20], and skin [21]. As a result of the pilot study for the GTEx tissue collection study, multiple studies involving genomic expression in esophageal carcinoma, lung adenocarcinoma, and breast tumors have been conducted (Figure 2 and Tables 2 and 3) [1, 2, 3, 18, 21].
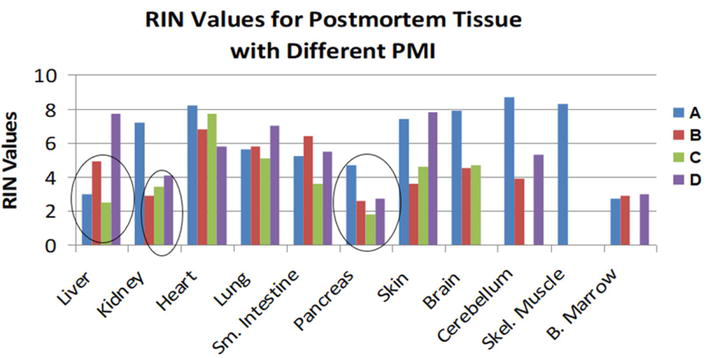
Figure 2.
Patients A-D RIN values for each tissue collected [
| Patient | A | B | C | D |
|---|---|---|---|---|
| Refrigeration (h) | 4.80 | 2.10 | 9.80 | 0.13 |
| Refrigeration Time (h) | 23.55 | 14.50 | 2.17 | 9.17 |
| Refrigeration to Autopsy (h) | 0.33 | 0.25 | 0.23 | 0.25 |
| Total time (h) | 28.68 | 16.85 | 12.20 | 9.55 |
Table 2.
Comparison of patients A-D PMI intervals [18].
| Patient | Agonal score | Agonal factors |
|---|---|---|
| A | 3 | Failed bone marrow transplant for multiple myeloma |
| B | 1 | CO2 Poisoning |
| C | 2 | Wound (more detail later) |
| D | 3 | 48 h medically-induced hypothermia during brain death protocol |
Table 3.
Agonal state score and factors of patients A-D [18].
4. Methodological considerations of brain and blood recovery, analysis, and storage
4.1 Introduction
The proper methodology for recovery, analyzing, and storage of human specimens is a common source of discussion among many scientists and researchers across the world. Many intrinsic and extrinsic factors such as temperature, timeliness of collection, and natural disease processes influence the quality of tissue, and thus the effectiveness to which the specimen can be analyzed. This section will discuss our SOP for brain banking and biological fluid in this brain collection. In general, our process includes protocols for the recovery, analysis, and storage of sections and entire brain specimens.
4.2 Methods used
Three main methods of collection in the suicidality project were used: blood collection, DNA, and RNA tubes. All methods were designed to allow an easy medium to which DNA and/or RNA may be extracted and analyzed. Blood was generally collected in PAXgene
The SOP began with deputy coroners/hospital staff notifying staff of a death that fits collection criteria guidelines. A taped verbal consent was obtained from the next of kin by action of the Deputy coroner and Followed up with obtaining signed written informed consent at the morgue or at the funeral home by the Coroner’s Office. Once these two actions were completed, the case was approved for tissue and/or blood collection.
The reagents include 4 mL labeled and bar-coded tubes for storage via PAX Gene tubes for RNA and DNA. If possible, whole blood was collected from normal patients and patients with neurological diseases. Tissue procurement included collection of PAXgene
Prolonged ischemic state of brain tissue, natural disease processes such as Alzheimer’s disease, and long postmortem intervals may have affected the quality of recovery of brain tissue and RIN values. While pH premortem acidosis often influenced viable brain tissue for genomic studies, pH is stable post-mortem and during consequent freezer storage. Typically, individuals who suffered prolonged agonal states tended to have more acidotic brains. Conversely, those who experienced brief deaths typically had a normal pH. RIN values in most postmortem tissues is highly correlated with medical history and agonal state. Natural disease processes and length of time taken to collect specimens after the decedent’s passing are taken into consideration. Special consideration was taken for subjects with a history of psychiatric disorders, psychiatric visits and hospitalizations, suicide attempts, and excessive use of alcohol and/or street drugs.
The inclusion and exclusion criteria were then further examined. Inclusion criteria consisted of the decedent’s medical history being consistent with schizophrenia, schizoaffective disorder, bipolar disorder, major depression, a postmortem interval of 24 h or less, any gender or race, 18 years or older (unless otherwise agreed upon in writing by a legal representative of the decedent), and appropriate informed consent forms. Oppositely, the exclusion criteria included greater than a 24 h post mortem interval, any primary or metastatic brain cancer, history of stroke, intracranial bleed or other brain trauma, severe alcoholism or Korsakoff’s syndrome, history of congenital brain defects, sepsis, infection, communicable disease, prolonged hypoxia or intubation/ventilator assisted breathing for greater than 12 hs, use of vasopressors prior to death, and attempted ACLS resuscitation.
Staff also developed strict quality control criteria. Upon each new sample obtained, the following quality control checks are performed: RIN ≥ 7.0 for frozen specimens, pH ≥ 6, proper FFPE specimen fixation, and appropriate preparation, labeling, and storage of samples, specimens, and data [19].
4.3 Concern for biohazard material
All human specimens should be considered potential biohazards whether they are fixed, paraffin-embedded, or fresh frozen. Due to the hazardous nature of the organs under examination, universal precaution was taken and standard procedures were implemented while handling the specimens.
4.4 Collection of brain
In addition to removal of the brain, factors of concern included the weight of the whole brain, bleeding, prior healed or open/partly open surgical incisions of the scalp or skull bone, dura stuck inside the calvarium or if it peeled away from the skull and stuck on the brain, and whether the surface of the meninges were stained with blood or were transparent. If any of these conditions were observed, the location and severeness of the afflictions were noted and photographed for documentation purposes.
The entire brain, including the cerebrum, cerebellum, and brain stem, are removed per standard autopsy procedure (Figures 3 and 4). The brain was then placed in a plastic bag partially filled with saline and placed in an ice-filled cooler. Upon arrival at the designated grossing area, the brain was divided by a mid-sagittal cut through the corpus callosum. A coronal sectioning was also performed through the brain hemisphere from anterior to posterior.
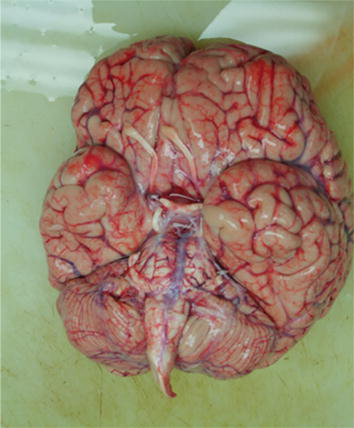
Figure 3.
Whole brain removed after prosection.
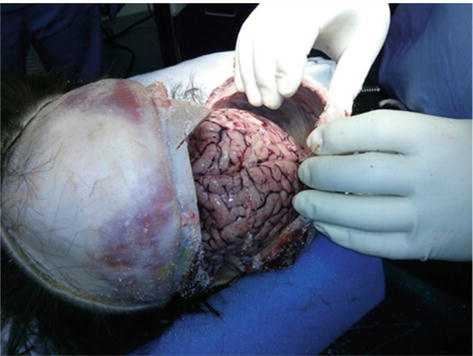
Figure 4.
Incision from ear to ear of decedent is then reflected in order to remove calvarium. Temporalis muscles removed in order for autopsy bone saw to properly cut through calvarium.
Next, a section of the identified region is cut that is approximately 1 cm thick. The section is then trisected to create three samples that are approximately 3 mm thick. Samples in the cassettes are then transported on wet ice and immediately (within 15 min) placed in a − 80°C freezer. The time that each sample is frozen is recorded. The last remaining sample is then placed into a white cassette and immediately placed in 10% neutral buffered formalin.
4.5 Storage of brain tissue
A pH strip was placed on the first section (about 10 seconds) to get the pH of the brain. Subsequent slices were made parallel to the first cut, producing 14 slices. The slices were then placed on a blue cutting board and photographed with a digital camera. Slices were slowly frozen using cooled stainless-steel plates. The precooled plates were then placed on ice packs in a flat Styrofoam container. Slight pressure on the upper stainless-steel plate allowed the cut brain slices to flatten out. More ice packs were then placed on top of the upper stainless-steel plate.
4.6 Conclusion
The data pertaining to all cases were compiled in a single database spreadsheet. Also present at each site for procurement, analysis, and storage are data books used to update the research study with pertinent findings and basic demographics. There were 180 total cases collected under the suicidality protocol and we had high-quality RNA data from blood samples. After seven brains were collected, we compared RNA profile from both brain and blood. Past findings showed the mutated genes were similar. The brain collection was stopped after the seventh case and are now only collecting DNA and RNA blood samples in PAXgene
5. Collection of lung and blood for COVID-19 post-mortem study
5.1 Introduction
In the past 3 years, a novel acute respiratory RNA virus known as SARS-Cov-2 has grown into a pandemic. In this investigation, the lung pathology of 43 individuals who passed away and displayed significant COVID-19 symptoms was examined. Decedents were not seen in a clinical environment and were taken post-mortem. Decedents ranged in age from 3 to 76 years old. Ethnicities included 29 Caucasians, eight African American, three Hispanic, one Cuban, one Asian, and one unknown. There was a male to female ratio of 4:1. All cases were classified into positive and negative COVID-19 using the FDA and CDC approved RT-PCR test. Eighteen were negative and twenty-one were positive for COVID. A subset of these cases (11) was positive for cocaine, methamphetamine, fentanyl, morphine, acetyl fentanyl, chronic ethanol, or sertraline. There were additional compounding etiologies with most of these cases (diabetes, trauma, and atherosclerotic heart disease) [22].
5.2 Concern for biohazard material
All human specimens should be considered as potential biohazards whether they are fixed, paraffin-embedded, or fresh frozen. Due to the hazardous nature of the organs under examination, universal precaution standard procedures were implemented while handling the specimens.
5.3 Tissue specimens
Informed consent and IRB approval was obtained. All lung and blood samples were collected at the Marion County Coroner’s Office. These were partial intercostal surgeries with a biological fluid blood draw over the period of April 9, 2020 to March 15, 2021.
Body transport: The body was transported from the scene of death to Marion County Morgue. The group used Tyvek Iso Clean surgical gowns over scrub suits, thick rubber gloves, surgical masks, eye goggle protection, surgical boots, and surgical slippers.
Lung removal: The clavicle was identified (rib 1 beneath clavicle) and counted down to ribs 3–5. An incision was made through the skin and through the thoracic cage from the mid-axillary line over to the manubrium of the sternum between ribs 4–7. This incision was approximately 10–12 cm in length. A vacuum was used to remove any fluid from the chest cavity. A small piece of lung tissue from the right middle lobe (5 cm × 7 cm) was removed and placed in 10% NBF. PAXgene
For this infectious disease lung study, lung tissues were fixed at 4°C for 48 h, instead of the usual 18 h followed by tissue processing, embedding in paraffin, and staining for H&E and Trichrome. Approximately 14 sections of lung were collected from each case.
Immunology: Antibodies were tested in various cases. Total RNA isolation was performed on all samples.
RNA extraction: RNA QC and qRT-PCR were used to confirm viral load in all 23 positive deceased blood and lung samples. The average post-mortem interval for these samples was 14 ½ h.
5.4 Findings
The findings were similar to previous COVID-19 findings. We had both acute and chronic inflammation in the lung samples. The histology in the acute cases were characterized by minimal bronchiolar perivascular lymphocytic infiltration, focal hyaline membrane formation, severe pulmonary congestion, bronchiolar respiratory epithelial/alveolar pneumocyte necrosis, formation of microthrombi, and pulmonary edema. Interstitial pneumonia with thickening of the alveolar wall, perivascular and peribronchiolar lymphocytic infiltration, multifocal areas of fibroplasia, microthrombi, severe congestion, squamous metaplasia of the alveolar epithelium, and fibrosis and septal collagen deposition were the hallmarks of the histology in the chronic cases.
We performed immunohistochemistry using both DNA and fluorescence imaging of lung tissue and detection of the COVID-19 virus. In addition, total RNA isolation with qRT-PCR and RNA Seq was completed to confirm the diseased blood in lung samples.
6. Collection of liver and lung for small cell lung cancer study
6.1 Introduction
This study examined a rapid autopsy program including three patients currently looking for mutational and molecular changes in small cell lung cancer (SCLC). The findings will provide new insights into treatment resistance and lead to new targets to find more effective immunotherapy and targeted therapies for advanced SCLC. SCLC is one of the rarest types of lung cancer, affecting 10–15% of all patients with lung cancer. Diagnosis usually occurs once the patient is symptomatic and after metastasis has occurred. It has one of the worst prognoses out of all lung cancers and is the hardest to treat. Blood samples from the superior vena cava were collected in the funeral home in order to keep the PMI under 8 h. Two more cases are currently being processed using this protocol. Most autopsy rooms do not collect tissue after usual business hours, and we have done all collections in a funeral home.
6.2 Specimen collection
Blood collection was performed utilizing the subclavian vein. Six tubes were collected: DNA, RNA, Peripheral blood mononuclear cells (PBMCs), Natural Killer (NK), circulating tumor cells (CTC), and In Vivo were collected (Figure 4). PAXgene
MRIs were performed to determine an appropriate location for lung and liver collection. Lung removal was characterized by an incision of 10–12 cm made through the skin through the mid-axillary line over to the manubrium of the sternum between ribs 3–7. Rib retractors were used to spread the ribs and have a large field to visualize the lesions in the lungs (Figure 5). Russian tissue forceps were used to remove the mass and adjacent normal tissue. A scalpel and/or autopsy knife is then used to dissect mass into 250–750 mg sample sizes. Masses were transferred to 10% neutral buffered formalin (NBF) and MACS solution.
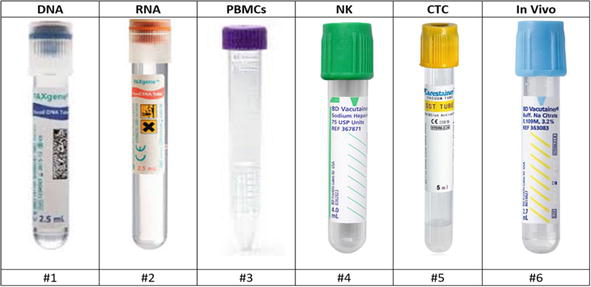
Figure 5.
Visual illustration of order of collection of blood tubes.
Liver removal was characterized by an incision of 10–12 cm made below the xiphoid process. The skin and fascia were lifted utilizing Russian tissue forceps, and a scalpel was used to cut into fascia. Liver palpation was performed to identify the mass and was carefully cut along with normal adjacent tissue. A vacuum to remove excess fluid accumulation from the abdominal cavity was used if necessary. Samples sizes of 250–750 mg were transferred into 10% NBF and transported on wet ice.
6.3 Subclavian vein location
The following anatomical landmarks were identified. The sternocleidomastoid muscle insertion into the clavicle was visible. Below the clavicle is the right subclavian vein. At the top of the ribcage, the right subclavian vein starts. The skin puncture site can be identified by landmarks 1 cm laterally from the sternocleidomastoid muscle’s posterior clavicular head and 1 cm down under from the clavicle (Figure 6).

Figure 6.
Rib spreader.
7. Collection of skin specimen
7.1 Introduction
The Post-mortem Tissue Collection Skin Project has collected 80 cases to date. Tissue collection was completed by the Marion County Coroner’s Office, and from there was transported on wet ice to the regenerative medicine lab where the skin samples were fixed and embedded into paraffin blocks. Skin samples were collected and divided into two samples. One went into 10% NBF and the other into MACS solution. MACS solution was kept in wet ice as it was transferred to the research lab. Microscope slides were prepared and stained with hematoxylin and eosin (H&E) and Masson’s Trichrome for autolysis detection. In addition, Visium spatial gene expression slides were created for molecular profiling. Tissues were profiled on a 4-point Hardy Score (Figures 7 and 8).

Figure 7.
Description of 4-point hardy scale.
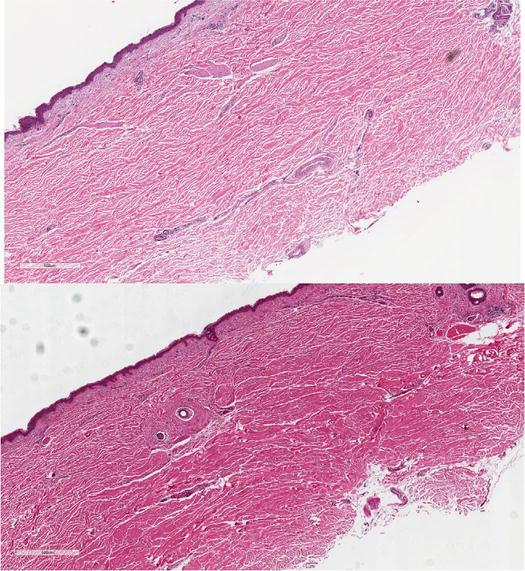
Figure 8.
Skin collected in MACS solution (top) compared to PAXgene® tissue fix (bottom).
7.2 Specimen collection
Skin specimens are identified with an alpha-numeric Specimen ID that contains the case ID and a sequence number. Collection time should stay under 16 h. A sun-exposed (e.g., lower limbs) 6x3 cm strip of skin is collected and cut into two sections. The tissue is preserved in MACS solution and PAXgene
7.3 Results
MACS solution separated the dermal collagen fibers in the dermis compared to the PAXgene
8. Conclusions
In recent years, the development of new molecular technologies with improved specificity, sensitivity, and throughput has greatly increased the demand for high quality, well-annotated biological specimens. By using surgical techniques instead of traditional autopsy protocols, we were able to maintain a short PMI and high genomic quality for samples collected. We have been able to collect tissues such as skin, lungs, and blood, advancing the research of post-mortem human genomics.
Acknowledgments
Thanks to Mikayla Stephens, Sarah Seibert, Kayli Nguyen, Maggie Granatir, Jessica Jackson, Matt Rodgers, Frank Lloyd, Dean Hawley, Rebekah Addison, Madelynn Gaines, Kanhaiya Singh, Chandan Sen, Charles Saller, and Raymond Howanski. We appreciate everyone who took the time to contribute to this research and the Marion County Coroner’s Office.
References
- 1.
Cancer Genome Atlas Research Network…Sandusky GE. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017; 541 (7636):169-175. DOI: 10.1038/nature20805 - 2.
Cancer Genome Atlas Research Network…Sandusky GE. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014; 511 (7511):543-550. DOI: 10.1038/nature13385 - 3.
Sandusky GE, Offen JC, Clark MA. A survey of human myocarditis cases diagnosed using Alexa immunofluorescence dyes and confocal microscopy. Life Science Journal. 2006; 3 (4):37-41 - 4.
Spencer C, Carr KA, Mitchell C, Sandusky GE. Quality control from a subset of human surgical tissue specimens from the IU Simon Cancer Center Tissue Bank collected in 2009-2010. In: An H&E and RIN Value Assessment: ISBER Annual Mtg; May 2011; Washington, DC. 2011 - 5.
Sandusky GE, Clark MA. Heart Transplantation and Evaluation of the Heart Explant. Madison, Wisconsin: University of Wisconsin; 2007 - 6.
Sandusky GE, Clark MA. Hypertrophic Cardiomyopathy: Midwest Pathology. Columbus, Ohio: Ohio State University; 2008 - 7.
Sandusky GE, Clark MA. Idiopathic Cardiomyopathy: Midwest Pathology. West Lafayette, IN: Purdue University; 2009 - 8.
Stan AD, Ghose S, Gao X, Roberts RC, Lewis-Amezcua K, Hatanpaa KJ, et al. Human post-mortem tissue: What quality markers matter? Brain Research. 2006; 1123 (1):1-11. DOI: 10.1016/j.brainres.2006.09.025 - 9.
Preece P, Cairns NJ. Quantifying mRNA in post-mortem human brain: Influence of gender, age at death, post-mortem interval, brain pH, agonal state and inter-lobe mRNA variance. Brain Research. Molecular Brain Research. 2003; 118 (1-2):60-71. DOI: 10.1016/s0169-328x(03)00337-1 - 10.
Hynd MR, Lewohl JM, Scott HL, Dodd PR. Biochemical and molecular studies using human autopsy brain tissue. Journal of Neurochemistry. 2003; 85 (3):543-562. DOI: 10.1046/j.1471-4159.2003.01747.x - 11.
International Society for Biological and Environmental Repositories. Best Practices: Recommendations for Repositories. 4th ed. Biopreserv Biobank; 2018 - 12.
National Cancer Institute. Best Practices for Biospecimen Resources. U.S. Department of Health and Human Services, National Institutes of Health. 2016 - 13.
Sandusky GE, Teheny KH, Esterman M, Hanson J, Williams SD. Quality control of human tissues--experience from the Indiana University Cancer Center-Lilly Research Labs human tissue bank. Cell and Tissue Banking. 2007; 8 (4):287-295. Epub 2007 Mar 27 - 14.
Catts VS, Catts SV, Fernandez HR, Taylor JM, Coulson EJ, Lutze-Mann LH. A microarray study of post-mortem mRNA degradation in mouse brain tissue. Brain Research. Molecular Brain Research. 2005; 138 (2):164-177. DOI: 10.1016/j.molbrainres.2005.04.017 - 15.
Cummings TJ, Strum JC, Yoon LW, Szymanski MH, Hulette CM. Recovery and expression of messenger RNA from post-mortem human brain tissue. Modern Pathology. 2001; 14 (11):1157-1161. DOI: 10.1038/modpathol.3880451 - 16.
Lipska BK, Deep-Soboslay A, Weickert CS, Hyde TM, Martin CE, Herman MM, et al. Critical factors in gene expression in post-mortem human brain: Focus on studies in schizophrenia. Biological Psychiatry. 2006; 60 (6):650-658. DOI: 10.1016/j.biopsych.2006.06.019 - 17.
Lee J, Hever A, Willhite D, Zlotnik A, Hevezi P. Effects of RNA degradation on gene expression analysis of human post-mortem tissues. The FASEB Journal. 2005; 19 (10):1356-1358. DOI: 10.1096/fj.04-3552fje - 18.
Szymanski M, Hawley D, Sandusky GE. Investigating the correlation between post-mortem interval and RIN values: A pilot study. International Journal of Nursing Research. 2023; 6 :1444. DOI: 10.29011/2688-9501.101444 - 19.
Le-Niculescu H, Levey DF, Ayalew M, Palmer L, Gavrin LM, Jain N, et al. Discovery and validation of blood biomarkers for suicidality. Molecular Psychiatry. 2013; 18 (12):1249-1264. DOI: 10.1038/mp.2013.95 - 20.
Abouhashem AS, Singh K, Srivastava R, Liu S, Steiner S, Gu X, et al. The prolonged terminal phase of human life induces survival response in the skin transcriptome. BioRxiv. 2023. DOI: 10.1101%2F2023.05.15.540715 - 21.
The GTEx Consortium. The genotype-tissue expression (GTEx) project. Nature Genetics. 2013; 45 (6):580-585. DOI: 10.1038/ng.2653 - 22.
Bloom A, Colter A, Jacobsen M, Battles D, Albertson T, Sandusky G. Pathological lung lesions of COVID-19 and opioid use disorder: A post-mortem lung and toxicology examination of 43 decedents. FMAR. 2022; 10 (04):65-76. DOI: 10.4236/fmar.2022.104007