Rapid antigen testing and sensitivity and specificity (adapted from Peeling [6]).
Abstract
Laboratory real-time qPCR (RT-qPCR) diagnosis and rapid antigen testing of COVID-19 variants have become a cornerstone of diagnosis of SARS-CoV-2 nucleic acids and antigens. This article proposes a comparative analysis of the benefits and limitations of these qualitative and quantitative methods through a literature review, and discusses how the validation of biomarker discovery in precision medicine could be applied to rapid antigen testing and molecular diagnostic workflows taking into considering testing sensitivity and specificity. Considerations of analytical validity and clinical validity are a focus. Diagnostic accuracy as shown by overall sensitivity and specificity of laboratory diagnostic RT-qPCR as compared with rapid antigen testing will be presented. This review is timely since the existing literature on RT-qPCR and rapid antigen testing for COVID-19 is significant containing large amounts of data, which at times is conflicting along with recommendations for streamlining these distinct methods for diagnostic testing of COVID-1 based on symptomatic presentation, vaccination and contact status. Since many cases currently are long COVID syndrome, the timeliness of the review may be paramount for potential future public health emergencies, especially involving respiratory illnesses.
Keywords
- real-time qPCR
- rapid antigen testing
- sensitivity
- specificity
- biomarker discovery validation
1. Introduction
SAR-CoV-2 virus, the causative agent of COVID-19, led to a worldwide pandemic starting in 2020 and is still ongoing as result of novel variants. It is a positive-sense single stranded RNA enveloped virus that is responsible for causing a rapidly contagious respiratory illness through close human interaction. The pandemic as a result of the COVID-19 outbreak was declared on in March 2020 by the World Health Organization [1]. As of February 2023, worldwide incidence and mortality exceeded 600 million and 6 million respectively.
The viral genome and structure includes two ORF (1a and 1b) genes that code for RNA-dependent RNA polymerase, helicase and protease and four structural proteins including the nucleocapsid N, membrane N, envelope E and spike glycoprotein surface S. The RNA polymerase, ORF genes, E, N and S genes serve as targets for molecular testing through the RT-qPCR technique (Figure 1) [2, 3].
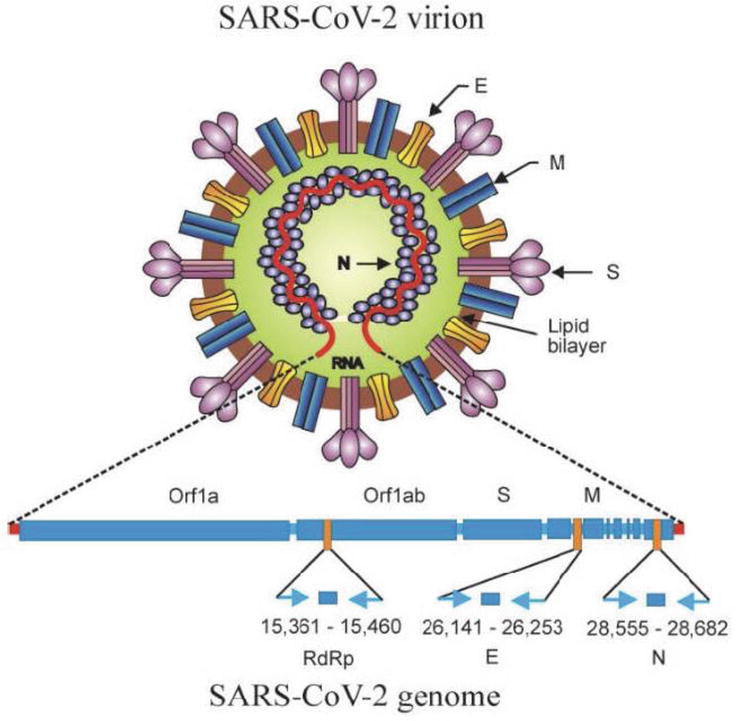
Figure 1.
Orf1a and Orf1ab and S and M genes (adapted from [
The functional receptor expressed on pulmonary epithelial cells, ACE-2 allows for the S protein to invade the host cell. The S protein is activated by cleavage, which leads to viral and host cell membrane fusion. The viral contents are emptied into the pulmonary alveolar epithelial cells and then the virus undergoes replication and the negative strand RNA is formed through RNA polyermase. The formed RNA strand serves as a template for the formation of positive RNAs which synthesize viral proteins in the cell cytoplasm, leading ultimately to viral particles that invade adjacent epithelial cells and potential infection via respiratory droplets through public transmission [4].
Efforts were directed to finding methods for diagnosis through tests that considered the virus’ properties and its genome and structure. For acute infection, nucleic acid detection (RNA) became the gold standard for revealing infection. Literature on standard RT-PCR testing found that these tests were sensitive and specific but also time-consuming, which led to finding alternative approaches for timely, cost-effective in-house diagnostic kits based on antigen assays detecting viral antigen. A meta-analysis conducted in 2020 found variance in the accuracy of quantitative testing according to prevalence of COVID-19, with post-test probabilities decreasing according to prevalence (50%: 96%; 20%: 84%; 55%: 5%). Subsequent meta-analysis showed increased testing accuracy however the variation still persisted. According to Menesez, a number of factors in SARS-CoV-2 detection that involve preanalytical and analytical aspects of detection, such as delays, lack of standardization in specimen collection, assays that have not been adequately validated and poor storage conditions prior to arrival result in failure in testing accuracy. This is compounded by potential for procedure contamination, the disease period incubation and viral mutations [5]. According to Peeling et al., “a [rapid antigen] test should be specific enough to minimize the proportion of cases erroneously diagnosed as positive in low prevalence settings, and sensitive enough to avoid missing a diagnosis as COVID-19 prevalence increases” [6].
This chapter conducts a review of COVID-19 rapid antigen tests and real-time PCR amplification assays in terms of their sensitivity and specificity in the literature and compares the data, and provides the published recommendations in determining the appropriate testing according to clinical presentation and contact status in the form of algorithmic workflows. Currently, many commercial SARS-CoV-2 detection kits serve as the reference standards. This chapter also proposes the application of biomarker discovery validation in precision medicine to COVID-19 laboratory molecular testing and rapid antigen testing.
2. Sensitivity and specificity for COVID-19 RT-PCR and antigen tests
The analytical validity of a diagnostic test is determined by the specificity and sensitivity of the assay. True positives are positives with the virus present; true negatives are negatives with the virus not present; false negatives are negatives with the virus present; false positives are positives without the virus present. Analytical validity and its referent to the rate of FP is seen the PPV of the test which mathematically is the PPV = TP/(TP + FP) x 100 and is the chance of disease upon receipt of a positive test and reflects disease prevalence, and specificity and sensitivity of the test [6].
2.1 Rapid antigen testing in asymptomatic and symptomatic COVID-19 patients: Emergency use authorization
A particle, fragment or molecule is considered an antigen whereby the immune system is triggered to produce antibodies to kill the pathogen. Rapid antigen testing in this context detects viral proteins, such as the S glycoprotein, M protein and the released N protein obviating the need for thermal amplification [1]. They reveal active viral infection and since antigens precede antibody and are targeted approaches, they do not reflect the recovery portion. Lateral flow assays or LFAs are considered the most commonly used rapid antigen testing for detection. The World Health Organization (WHO) recommended a minimum sensitivity of 80% and a minimum specificity of 97% for effective utilization of rapid antigen testing [7].
The analytical parameters of the Roche SARS-CoV-2 Rapid Antigen Test was determined by assaying negative and positive PCR samples and according to one study. 75 nasopharyngeal swabs from patients which tested SARS-CoV-2 RNA-negative were tested with the Real Star SARS-CoV-2 RT-PCR Kit “were collected and investigated using the SARS-CoV-2 Rapid Antigen Test, a lateral flow assay.” The interpretation of lateral flow assays on samples showed that when both control line and test line were present, the test was regarded as SARS-CoV-2 antigen negative. When only the control line was present, the test was regarded as SARS-CoV-2 antigen negative. 75 positive nasopharyngeal swabs from patients that demonstrated huge variation in cycle thresholds and tested SARS-CoV-2 RNA positive with the Real Star SARS-COV-2 RT-PCR Kit were investigated using the SARS-CoV-2 Rapid Antigen Test cycle (Ct < 20: n = 5, Ct 20- < 25: n = 12, Ct25- < 30: n = 20, Ct 30- < 35: n = 29, Ct > = 35: n = 9). The specificity was determined as 96% and assay’s “sensitivity with samples with a cycle threshold of < 25, 25 - <30, 30 - <35, and> = 35 was 100 %, 95 %, 44.8 % and 22.2 %, respectively.” The authors note that these values are inferior to the PCR assay in terms of sensitivity and specificity but conclude that this alternative rapid antigen test is easier and more convenient to distinguish contagious from non-contagious individuals [8].
In 2021, a retrospective, cross-sectional study of an asymptomatic student population called StudyCov (n = 692) analyzed the specificity of the RT-PCR BioMerieux SAR-CoV 2 antigenic test. “The index test was the nasopharyngeal Abbott Panbio SARS-CoV-2 Ag rapid test” When stratified by symptoms and risk exposure, StudyCov revealed the following results:
Overall sensitivity and specificity:
65% [95%CI:49.0–76.4%]
100% [95%CI:99.4–100%] respectively.
In the asymptomatic sub-group:
35% [95%CI: 15.4–59.2%] and 100% [95%CI: 99.3–100%] respectively.
“Using AT instead of RT-PCR, 13 (65%) subjects would have been missed, including 10 considered likely contagious.”
The authors conclude: “This study shows the poor sensitivity of RAT (rapid antigen testing) in asymptomatic subjects, specificity being however excellent. The performance results fall below the World Health Organization recommendation of 80% sensitivity and question using AT in general population, especially when asymptomatic” [7].
Additional studies revealed the sensitivity and specificity of the Abbott BinaxNOW and Panbio test in asymptomatic patients. The evaluation of Abbott BinaxNow was conducted in asymptomatic walk-up patients and compared to RT-PCR: 3% had a positive RT-PCR. Sensitivity and specificity were respectively 93.3% (95% CI: 68.1–99.8%) and 99.9% (95% CI: 99.4–99.9%) for a Ct < 30 case definition. Unlike StudyCov, “the swab was done in both nostrils for both tests, the population was older and the AT was different,” and “those differences might partly explain the performance differences”. The Panbio SARS-CoV-2 AG Rapid Test Device with RT-PCR for emergency units-referred patients, 72.1% of which were asymptomatic. Sensitivity was 73.3% (95% IC: 62.2–83.8%) which is consistent with the BioMerieux results [9].
One field study conducted a rapid antigen detection test on infected individuals who were asymptomatic in an evaluation of the Panbio COVID Nasopharyngeal swabs were collected from household and non-household contacts. Confirmatory testing was done with the TaqPath Combo Kit using RT-PCR. They found that of 79 individuals tested positive by RT-PCR of whom 38 yielded positive RAD (rapid antigen detection) results. The overall sensitivity and specificity was 48.1% (95% CI 37.4–58.9) and 100%(95% CI 99.3–100), respectively, with sensitivity being higher in household (50.8%) than non-household contacts (35.7%); symptomatology presentation was more probable in RAT positive patients (p < 0.001). They conclude that “the Panbio test displays low sensitivity in asymptomatic close contacts of COVID-19 patients, particularly in non-household contacts. Nonetheless, establishing the optimal timing for upper respiratory tract collection in this group seems imperative to pinpoint test sensitivity” [9].
Table 1 shows the list as of September 2021 as compiled by Peeling et al. of rapid antigen tests and their sensitivities and specificities [6].
| Sample type | Time of sample collection* | Result reading | Sensitivity specificity† | Comments | |
|---|---|---|---|---|---|
| Abbott BinaxNOW, USA | Nasal swab | 0–7 days | Visual, 15 min | 97%, 99% | WHO Emergency Use Listing; US FDA Emergency Use Authorization; app for results; influenza A and B tests available |
| Abbott Panbio, USA | Nasal swab, nasopharyngeal swab | 0–7 days | Visual, 15–20 min | 93%, 99% | WHO Emergency Use Listing, US FDA Emergency Use Authorization pending |
| Access Bio CareStart, USA | Nasal swab, nasopharyngeal swab | 0–5 days | Visual, 15-20 min | 88%, 100% | US FDA Emergency Use Authorization |
| BD Veritor, USA | Nasal swab | 0–5 days | Instrument, 30 min | 84%, 100% | US FDA Emergency Use Authorization |
| LumiraDx, UK | Nasal swab | 0–12 days | Instrument, 12 min | 98%, 97% | US FDA Emergency Use Authorization |
| Quidel Sofia SARS Antigen Fluorescent Immunoassay, USA | Nasal swab nasopharyngeal swab | 0–5 days | Instrument, 20 min | 97%, 100% | US FDA Emergency Use Authorization; does not differentiate between SARS-CoV and SARS-CoV-2 |
| Quidel Sofia Flu and SARS Antigen Fluorescent Immunoassay, USA | Nasal swab, nasopharyngeal swab | 0–5 days | Instrument, 20 min | 95%, 100% | US FDA Emergency Use Authorization |
| SD Biosensor, South Korea | Nasal swab, nasopharyngeal swab | Not stated | visual, 15-30 min | 97%, 100% | WHO Emergency Use Listing |
Table 1.
Days after symptom onset.
Data from manufacturers.
Data from the Foundation for Innovative New Diagnostics.2 SARS-CoV = severe acute respiratory syndrome coronavirus. FDA = Food and Drug Administration.
2.2 Sensitivity and specificity of quantitative real-time PCR COVID-19 diagnostic testing
When the pandemic cases were increasingly rising in early 2020, the nucleic acid amplification test quantitative real-time PCR began to serve as a major platform for the diagnosis of SARS-CoV-2 viral RNA [2].
Real-time PCR is a quantitative test that uses the principles of PCR for fast, accurate detection of the amplicon through the use of fluorescent dyes such as SYBRGreen and probes such as TaqMan. Viral RNA is reverse transcribed into cDNA which is then converted by Taq polymerase into double stranded DNA. Target genes detected by the amplification process included the ORF1ab EN, ORF1ab N and ORF1 ab NS “Viral load is determined through a threshold indicated by Ct level and quantified by levels of double stranded DNA indicated by fluorescence intensity” (Figure 2) [1].
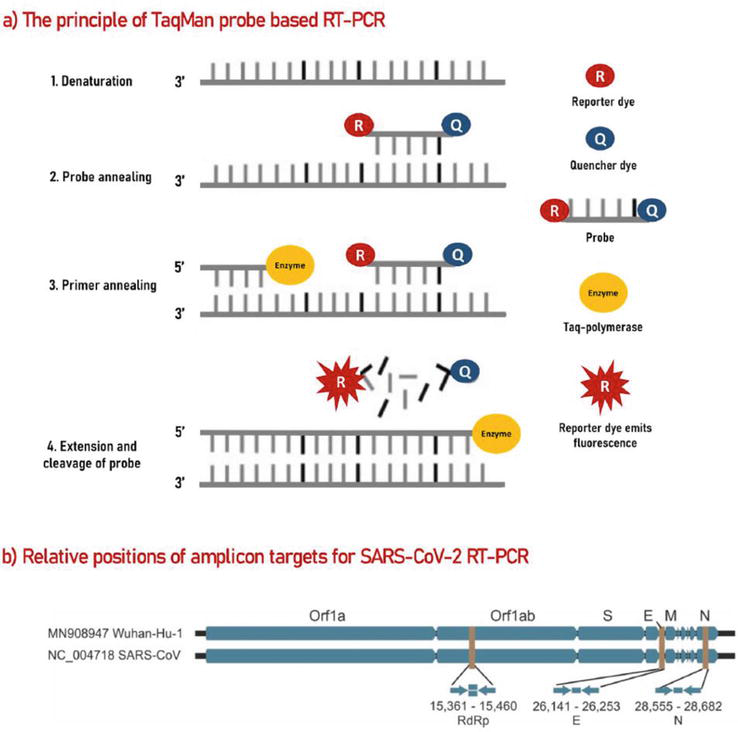
Figure 2.
Mechanism of quantitative RT-PCR method (adapted from [
When cases began to multiply at an increasingly alarming rate, real-time PCR emerged as the gold standard for diagnosis due to high sensitivity and specificity for symptomatic and asymptomatic carriers of infection. Banko et al. in [3] compared three real-time PCR tests and determined analytical sensitivity limit of detection at 500 copies/mL: The GeneFinder COVID-19 Plus RealAmp Kit (OSANG Healthcare Co., Anyang, Korea); Sansure Biotech (Sansure Biotech Inc., Changsha, China); TaqPath COVID-19 CE-IVD RT-PCR Kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA), which amplify the ORF1ab and N genes. Tables 2 and 3 show the positive and negative results out of 354 positive COVID-19 specimens. The authors conclude that there are similar analytical sensitivities and diagnostic accuracy. Final results indicated that the Sansure Biotech RT-qPCR method had significantly more positive results in comparison to the GeneFinder and TaqPath RT-qPCR methods (p < 0.001 and p < 0.001, respectively), and the GeneFinder RT-qPCR had significantly more positive results in comparison to the TaqPath RT-qPCR method (p 0.001 [3]).
| RT-qPCR Method | Target Gene | Positive, | Negative, |
|---|---|---|---|
| Only ORFlab | 4 (1.1) | 350 (98.9) | |
| Only N | 12 (3.4) | 342 (96.6) | |
| Both ORFlab and N | 190 (53.7) | 164 (46.3) | |
| 206 (58.2) | 148 (41.8) | ||
| Only ORFlab | 6 (1.7) | 348 (98.3) | |
| Only N | 11 (3.1) | 343 (96.9) | |
| Only E | 176 (49.7) | 178 (50.3) | |
| Both ORFlab and N | 176 (49.7) | 178 (50.3) | |
| 193 (54.5) | 161 (45.5) | ||
| Only ORFlab | 0 | 354 | |
| Only S | 0 | 354 | |
| Only N | 24 (6.8) | 330 (93.2) | |
| Both ORFlab and S | 0 | 354 | |
| Both ORFlab and N | 132 (37.3) | 222 (62.7) | |
| Both S and N | 0 | 354 (100) | |
| ORFlab and S and X | 22 (6.2) | 332 (93.8) | |
| 178 (50.3) | 176 (49.7) | ||
Table 2.
RT-PCR tests comparison (adapted from [3]).
| RT-qPCR Method | Measure of Diagnostic Accuracy with Its 95% CI | ||||||
|---|---|---|---|---|---|---|---|
| Sn (%) | Sp (%) | Overall Accuracy (%) | LR+ | LR– | PPV (%) | NPV (%) | |
| 99.5 (98.5–1.00) | 91.3 (87.0–95.7) | 95.8 (93.7–97.9) | 11.440 (6.94–18.87) | 0.006 (0.001–0.040) | 93.2 | 99.3 | |
| 99.4 (98.3–1.00) | 90.9 (86.7–95.2) | 95.2 (93.0–97.4) | 10.94 (6.86–17.45) | 0.006 (0.001–0.044) | 91.7 | 99.4 | |
| 99.4 (98.3–1.00) | 83.5 (78.0–89.0) | 91.5 (88.6–94.4) | 6.03 (4.33–8.42) | 0.01 (0.001–0.05) | 85.9 | 99.9 | |
Table 3.
Diagnositc accuracy of RT-PCR tests (adapted from [3]).
Teymouri et al. stated that “Unlike other molecular tests that do not have perfect diagnostic specificity, qRT-PCR is highly specific with a specificity of almost 100 %” and “has led to RT-PCR becoming the gold standard molecular diagnostic test.” However, pitfalls when the assay was being developed emerged in detecting true positives and true negatives in infected patients, and providers were counseled to combine molecular and clinical evidence. Sensitivity was reported at 45–60% in nasal swab samples and a false negative rate of 27% within the seven days when the illness onset in hospitalized patients. Patients diagnosed with chest CT scans and acute respiratory symptoms when molecularly tested on respiratory samples had a 33% false negative rate, as reported by researchers. This high false negative rate is attributed to variation in time of sampling since the onset of symptomatology, with values ranging from to 67% (day 1) to 38%, (symptom manifestation) and gradually returning to 66% on day 21 after symptoms have been observed (Table 4) [11].
| Name of the Kit | Target Genes | Type | Sample Preparation | No. of Tests | Time | LOD | Sensitivity | Specificity | Cost (Per Test) |
|---|---|---|---|---|---|---|---|---|---|
| CovidNudge | rdrp1, rdrp2, E gene, N gene, n1, n2, and n3 | RT-PCR | Automated | NA | ~90 min | 5 copies/μL | >94% | 100% | GBP 10 |
| Accula SARS-CoV-2 Test | N gene | RT-PCR | Automated | NA | ~30 min | NA | 100% | 100% | USD 20 |
| Cepheid Xpert Xpress SARS-CoV-2 assay | N2 and E | RT-PCR (real time) | Automated | 10 per kit | 0.02 PFU/mL | USD 19.8 | |||
| FastPlex Triplex SARS-CoV-2 Detection Kit | ORFlab, N, RPP30 | RT-dPCR | Manual | 96 test per kit | 90 min | 285.7 copies/mL | >95% | 95.7% | USD 1152 |
| Gnomegen COVID-19 RT-Digital PCR Detection Kit | N1, N2 | RT-dPCR | Manual | 48 samples per day | 180 min | 2.5 copies per reaction | >95% | 99% | NA |
| Bio-Rad SARS-CoV-2 ddPCR Test | N1, N2 | RT-dPCR | Manual | 96 samples | NA | 400 copies/mL | NA | ||
| ePlexSARS-CoV-2 Test | N gene | End-point RT-PCR with electrochemical Detection | Automated | 12 tests/kit | NA | 1 × 103 copies/mL | 99.02% | 98.41% | NA |
Table 4.
RT-PCR tests and sensitivity and specificity (adapted from [10]).
RT-PCR results interpretation for the COVID-19 RT-PCR Single Plex Test kit by LabCorp shown in Table 5 demonstrates that the test becomes “invalid” when the genes test negative. When the viral N genes test positive, the result displays “SARS-CoV-2 positive”. The result is interpreted as “intermediate” when one only one of the target viral genes tests positive, requiring another testing. When all viral N genes test negative, the test result becomes “SARS-CoV-2 negative” [1].
| SARS- CoV-2 N1 (FAM) | SARS- CoV-2 N2 (FAM) | SARS- CoV-2 N3 (FAM) | RNAse P (FAM) | Interpretation | Report | Actions (Clinical Site Samples) | Actions (Pixel Home Collection Kit samples) |
|---|---|---|---|---|---|---|---|
| + | + | + | +/− | SARS-CoV-2 detected | DETECTED | Report results to sender and appropriate public health authorities | Report results to PWN Health, who will call the patient. Report the result to the appropriate public health authorities |
| If only one target is positive | +/− | +/− | SARS-CoV-2 Indeterminate | INDETERMINATE | The sample is repeated once. If results remain the same, it is reported to the sender as indeterminate and recommend recollection if the patient is still clinically indicated | The sample is repeated once. If results remain the same, it is reported to the sender as indeterminate to PWN Health, who will call the patient. Report the result to the Pixel Portal | |
| — | — | — | + | SARS-CoV-2 Not Detected | NOT DETECTED | Report results to sender | Report result to PWN Health and the Pixel Portal |
| — | — | — | — | Invalid Result | INVALID | The sample is repeated once. If a second failure occurs, it is reported to the sender as invalid and recommend recollection if the patient is still clinically indicated | The sample is repeated once. If a second failure occurs, it is reported to PWN Health. Pixel’s customer service will contact the patient to discuss options. Report the result to the Pixel Portal |
Table 5.
Determination of validity of SARS-CoV-2 test (adapted from [1]).
2.3 Comparison and assessment of rapid antigen testing and real-time PCR
1000 types of molecular and antigen-based immunoassay tests are commercially available. According to one report, NAATs (nucleic acid amplification tests) such as RT-PCR can result in a positive test for weeks to months post infection, and may detect remnant viral RNA well after recovery. In contrast, antigen-based assays remain positive for 5–12 days subsequent to the onset of symptoms, and perform better in persons exhibiting a high viral load, which correlates with disease severity and death. Thus, rapid antigen testing may be a good indicator of potential transmissibility and correlate better than molecular tests with replication competent SARS-CoV-2 [12].
The rapid antigen tests are similar to PCR based tests in that detection is revealed during active viral infection rather than during the recovery situation. However, they are considered more reliable than antibody tests since they are target-specific. According to Yuce et al., “Antigen tests can be operated on LFA strip for rapid detection purposes or in ELISA format for better sensitivity, and high throughput uses (the simultaneous measurement of 96 samples).” A fluorescent immunochromatographic LFA assay for detecting the nucleocapsid (N) protein of SARS-CoV-2 was developed which “utilizes anti-N mouse antibodies and goat anti-rabbit IgG antibodies to create the test and control lines, respectively. It uses anti-N rabbit IgG marked with carboxylate-modified polystyrene Europium (III) chelate microparticles as signal particles” [1].
Repetitive RT-PCR testing has been shown to enhance performance characteristics, with false positive rates governed by differing factors, including time after onset of symptoms, symptom severity and nasal or oropharyngeal specimen used. Figure 3 shows the timeline of viremia, antigenic and immunogenic response, illustrating the timepoints when the rapid antigen testing and RT-PCR are most sensitive. Figure 4 illustrates a workflow whereby antigen and molecular testing can prove to be most accurate in the identification of the virus and resolution of symptoms, with different pathways for symptomatic and asymptomatic patients. When comparing the diagnostic tests, the predictive value of RT-PCR is higher and has higher post-test probability of reflecting the infected status of the individual, according to recent studies.
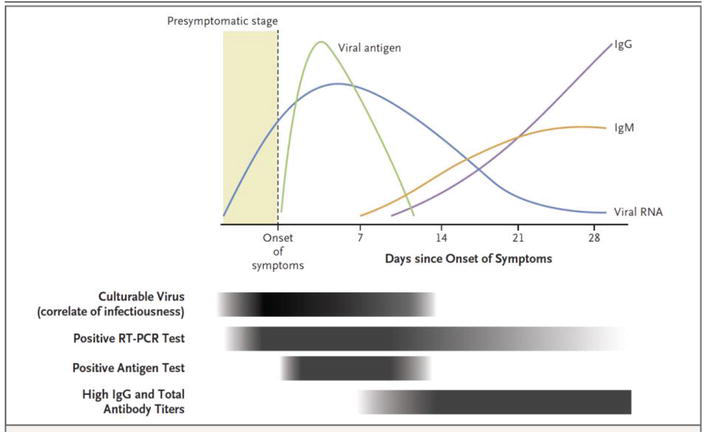
Figure 3.
Pathophysiology of viremia with positive molecular and rapid antigen tests output (adapted from [
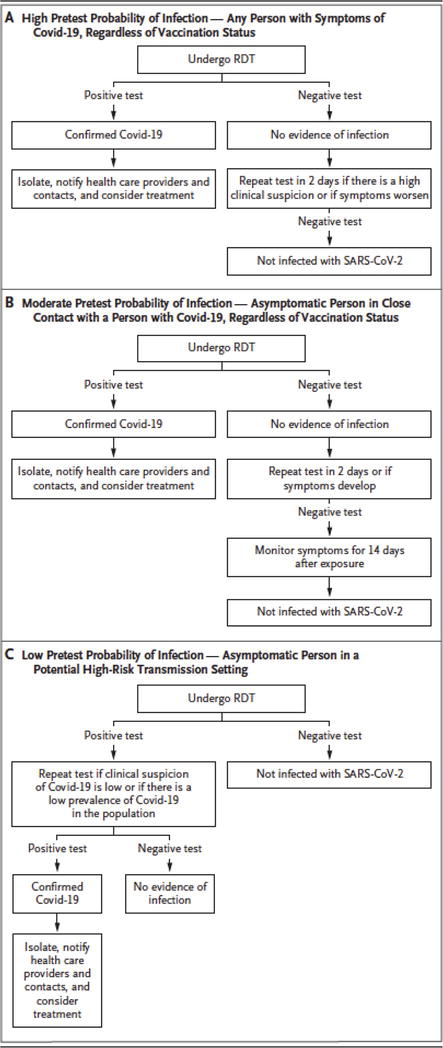
Figure 4.
Diagnostic testing workflows based on symptomatic status and patient management from diagnostic results (adapted from [
3. Discussion: biomarker discovery validation and COVID-19 diagnostic testing
Analytical validation of biomarkers for use in genomic testing were developed and optimized for the sequencing of somatic samples, which became central to the implementation of precision medicine [13]. This paradigm can be adapted for COVID-19 molecular and rapid antigen testing for the purposes of streamlining sensitivity and specificity data. Pre-analytical and analytical validation criteria for biomarker validation may be applicable for COVID-19 testing. Tables 6 is adapted from biomarker validation criteria and show how disease presentation and sample collection can lead to more accurate testing and better predictive value. The parameters in the second column show how assessment of pre-analytical characteristics, such as symptomatology, determination of test strategy, disease incidence and prevalence and accuracy in sample collection can form the basis of high analytical validity as determined by test choice of rapid antigen testing and compared with gold standard in asymptomatic and symptomatic individuals. Testing data generated can form the basis of high accuracy and evidence for analytical and clinical validation.
| Pre-Analytical Assessment |
|
| Analytical Validation |
|
Table 6.
Applying biomarker development validation guidelines to rapid antigen and molecular diagnostic testing (adapted from [13]).
4. Conclusion
While the WHO announced the sensitivity criteria for rapid antigen testing, molecular testing remains the reference standard, despite logistical difficulties in personnel and laboratory instrument maintenance. Several studies have been performed comparing the two methods resulting in patient workflows as shown in recent reports. False negatives and false positives complicate the interpretative power of these tests, since erroneous clinical decisions would ultimately impact public health outcomes. Biomarker analytical validation can serve as a foundation for further streamlining of sensitivity of these results and possibly lead to more propitious control of pandemics.
References
- 1.
Yuce M, Filizekin E, Ozkaya KG. COVID-19 diagnosis—A review of current methods. Biosensors and Bioelectronics. 2020; 172 :112752 - 2.
Artika IM, Dewi YP, Nainggolan IM, Siregar JE, Antonjaya U. Real-time polymerase chain reaction: Current techniques, applications, and role in COVID-19 diagnosis. Genes. 2022; 13 :2387 - 3.
Banko A, Petrovic G, Miljanovic D, Loncar A, Vukcevic M, Despot D, et al. Comparison and sensitivity evaluation of three different commercial real-time quantitative PCR kits for SARS-CoV-2 detection. Viruses. 2021; 13 :1321 - 4.
Parashar A. COVID-19: Current understanding of its pathophysiology, clinical presentation and treatment. Postgraduate Medical Journal. 2021; 97 :312-320 - 5.
Menezes S, Pestana DVS, Ferreira JC, Ribeiro de Carvalho CR, Felix MC, et al. Distinct outcomes in COVID-19 patients with positive or negative RT-PCR test. Viruses. 2022; 14 :175 - 6.
Peeling RW, Olliaro PL, Boeras DI, Fongwen N. Scaling up COVID-19 antigen tests: promises and challenges. The Lancet Infectious Diseases. 2022; 21 :e290-e295 - 7.
Ferte T, Ramel V, Cazanave C, Lafon M-E, Bebear C, et al. Accuracy of COVID-19 rapid antigenic tests compared to RT-PCR in a student population: The StudyCov study. Journal of Clinical Virology. 2021; 141 :104878 - 8.
Krüttgen A, Cornelissen CG, Dreher M, Hornef MW, Imohl M, Kleines M, et al. Comparison of the SARS-CoV-2 rapid antigen test to the real star Sars-CoV-2 RT PCR kit. Journal of Virological Methods. 2021; 288 :114024 - 9.
Torres I, Poujois S, Albert E, Colomina J, Navarro D. Evaluation of a rapid antigen test (Panbio™ COVID-19 Ag rapid test device) for SARS-CoV-2 detection in asymptomatic close contacts of COVID-19 patients. Clinical Microbiology and Infection. 2021; 27 :636.e1-636.e4 - 10.
Gupta N, Augustine S, Narayan T, O’Riordan A, Das A, et al. Point-of-Care PCR assays for COVid-19 detection. Biosensors. 2021; 11 :141 - 11.
Teymouri M, Mollazadeh S, Mortazavi H, Ghale-noie ZN, Keyvani V, et al. Recent advances and challenges of RT-PCR tests for the diagnosis of COVID-19. Pathology- Research and Practice. 2021; 221 :153443 - 12.
Drain P. Rapid diagnostic testing for SARS-CoV-2. New Eng. Journal of Medicine. 2022:1-9 - 13.
Hays P. Advancing Healthcare through Personalized Medicine. 2nd ed. Cham, Switzerland: Springer Nature; 2021